Abstract
Nutrient and energy sensing and signaling mechanisms constitute the most ancient and fundamental regulatory networks to control growth and development in all life forms. The target of rapamycin (TOR) protein kinase is modulated by diverse nutrient, energy, hormone and stress inputs and plays a central role in regulating cell proliferation, growth, metabolism and stress responses from yeasts to plants and animals. Recent chemical, genetic, genomic and metabolomic analyses have enabled significant progress toward molecular understanding of the TOR signaling network in multicellular plants. This review discusses the applications of new chemical tools to probe plant TOR functions and highlights recent findings and predictions on TOR-mediate biological processes. Special focus is placed on novel and evolutionarily conserved TOR kinase effectors as positive and negative signaling regulators that control transcription, translation and metabolism to support cell proliferation, growth and maintenance from embryogenesis to senescence in the plant system.
Introduction
The target of rapamycin (TOR) is an atypical serine-threonine protein kinase (PK) closely related to the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family conserved from yeasts to plants and humans. Extensive research over the past decade has demonstrated a pivotal role of TOR in sensing and responding to nutrient availability, cellular energy status, as well as stress and growth stimuli to drive cellular and organismal growth in all eukaryotes [1–5, 6*]. In photosynthetic organisms from unicellular Chlamydomonas reinhardtii to flowering plants, TOR has emerged as a central integrator of nutrient, energy and stress signaling networks [4,6*,7,8*]. Recent studies have also led to new links for TOR-regulated translational reinitiation of specific mRNAs stimulated by auxin [9**] and viral infection [10].
How TOR kinase modulates a broad spectrum of cellular processes from transcription, translation to metabolic reprogramming to support cell proliferation and growth has been the focus of intensive research. In the budding yeast Saccharomyces cerevisiae, Sch9 (an AGC family kinase and ortholog of plant and mammalian small ribosome protein S6 kinase, S6K), Tap42 (a regulator of PP2A phosphatases and ortholog of plant type 2Aphosphatae-accociated protein 46 kD, TAP46), and Atg1 (an ortholog of plant and mammalian autophagy related kinase, ATG1/ULK1) are three direct effectors of TOR complex 1 (TORC1) that act as master regulators of transcription, protein synthesis and autophagy [3,11,12]. More elaborate TOR signaling networks are emerging in multicellular animals and plants. Current knowledge indicates that mammalian TORC1 phosphorylates S6K, 4E-BP (eukaryotic translation initiation factor 4E binding protein), GRB10 (growth factor receptor-bound protein 10), LIPIN and ATG1/ULK1 to control directly translation and autophagy, but indirectly transcription [5]. Whereas mammalian TORC2 could phosphorylate AKT to regulate cytoskeleton structure, glycolysis, glycogenesis, and lipogenesis, there is no evidence to support the presence and function of TORC2 in plants yet [6*]. Although the phosphorylation and regulation of S6K, TAP46, LIPIN and ATG1 by plant TOR kinase may share some functional and mechanistic conservation as those in the budding yeast and mammals, recent discoveries have identified previously unknown TOR substrates and regulatory mechanisms in transcription, as well as ribosome biogenesis and translational controls critical to plant cell proliferation and growth regulation. Comprehensive review articles have summarized recent studies of Arabidopsis tor and related mutants [13**,14,15,16**], which unravel the multifaceted roles of TOR signaling in plant growth, metabolism and senescence [4, 6*, 17,18]. This review highlights the latest progress on applying chemical and genetic perturbations to identify novel molecular links and elucidate regulatory mechanisms in the plant TOR signaling network.
Exploring chemical tools to uncover diverse TOR functions
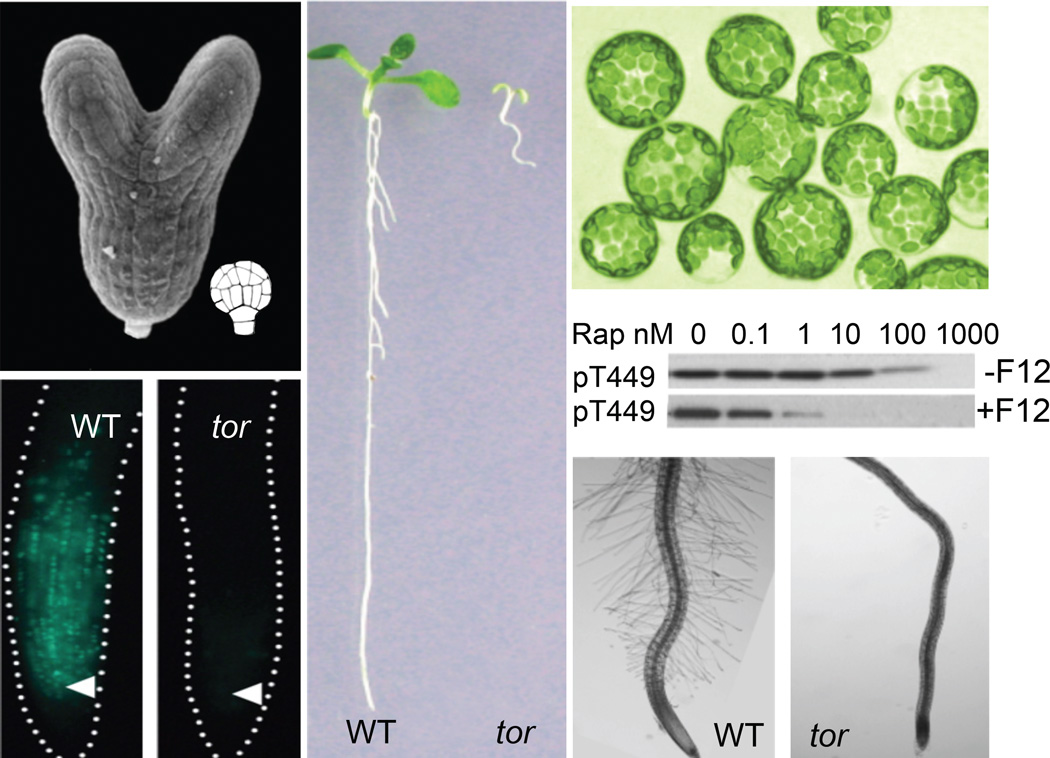
The characterization of many tor null mutants confirms that TOR deficiency results in early embryo arrest and lethality in Arabidopsis (Figure 1), which supports its essential role in plant growth and development but hindered the investigation of more TOR functions and regulatory mechanisms [19,20]. Although green alga C. reinhardtii is sensitive to rapamycin at 500 nM [21–23], early studies suggested that flowering plants were insensitive to rapamycin and the FKBP-rapamycin-binding domain (FRB) of Arabidopsis TOR did not interact with the Arabidopsis FKBP12-rapamycin complex in yeast two-hybrid and in vitro pull down analyses [19,24]. Unexpectedly, in vivo split luciferase protein interaction assay using Arabidopsis mesophyll protoplast revealed that Arabidopsis and human FKBP12 exhibited similar interactions with the FRB domain of Arabidopsis TOR (AtTOR) stimulated specifically by rapamycin [25]. The fully differentiated leaf mesophyll cells express the TOR protein and maintain robust endogenous TOR kinase activity for specific Arabidopsis S6K1 and S6K2 phosphorylation, which is sensitive to 100 nM rapamycin or even 1 nM rapamycin with FKPB12 overexpression (Figure 1). Therefore, in plant cells, AtTOR is sensitive to rapamycin within the range of the previously defined “physiologically” effective concentrations observed in yeast, C. reinhardtii and mammalian cells [21, 22, 25, 26*].
Figure 1.
Central roles of TOR in early embryogenesis, cell cycle control, and plant growth and development. The null tor mutants arrest at the 16–32 cell stage in early embryogenesis. Estradiol-inducible tor mutants block DNA synthesis in the root meristem, seedling development, and root hair growth. Fully differentiated leaf mesophyll cells maintain robust TOR kinase activity for S6K1-T449 phosphorylation that is sensitive to rapamycin without (upper panel) or with (lower panel) FKBP12 (F12) overexpression. Rap, rapamycin.
A previous study indicated that AtTOR is expressed in primary meristem, embryo and endosperm but not in differentiated cells [19]. A recent study shows that pTOR:GUS is ubiquitously expressed in the seedling and inflorescence tissues [15] and TOR-GFP is detected in the cytosol and nucleus [20]. Thus, TOR actions are not restricted to embryos, meristems and growing cells, or limited to translational control [27]. It is likely that TOR may have broad functions in regulating transcription, translation, bioenergetics, metabolism, stress and immune responses in diverse plant cell types, tissues and organs, which have not been yet fully recognized and investigated [6*,13**,14,15,16**,20,25]. Highly sensitive plant cell-based assays, liquid-medium seedlings more amenable to chemical treatment, as well as FKBP12 overexpression transgenic plants offer new opportunities for plant TOR research by overcoming ineffective rapamycin treatment and circumventing embryo lethality (Figure 1) [6*,15,16**,25].
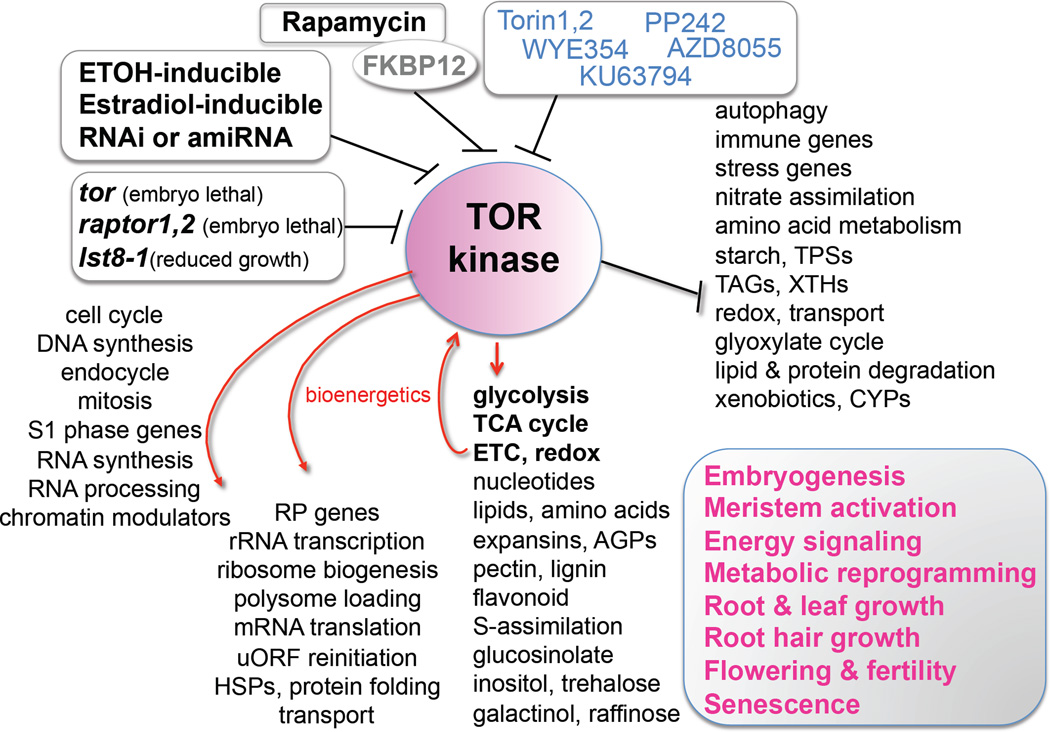
Besides the rapamycin-sensitive TOR signaling functions, molecular, genetic and biochemical studies have also identified rapamycin-insensitive TOR effectors that are directly but differentially phosphorylated by mammalian TOR kinase [26*]. Recently, a new generation of ATP-competitive chemical inhibitors specific to TOR kinase, such as Torin1, Torin2, WYE354, WYE132, KU63794, PP242 and AZD8055, have been described and readily available (Figure 2) [16**,28,29*]. These chemical inhibitors retarded root elongation in a TOR gene-dosage-dependent manner and blocked meristem growth, cell proliferation and root hair expansion in Arabidopsis seedlings, resembling the inducible null tor mutants (Figure 1) [16**,25,29*]. Similar chemical inhibitor doseresponses in root and root hair growth was observed in divergent angiosperms, including Nicotiana benthamiana, Lotus japonicus, Panicum miliaceum, and Oryza sativa [29*]. Future application of specific chemical inhibitors may enable the identification of both rapamycin-sensitive and rapamycin-insensitive effectors of TOR kinase, which will greatly expand our understanding of the regulatory mechanisms in the plant TOR signaling network. Although some of these chemical inhibitors may affect off-target PKs based on various mammalian kinome-wide assays, it is important to note that many of these mammalian off-target PKs do not exist in plants [28, 30]. The combinatorial application of multiple chemical inhibitors with specificity to TOR kinase but without overlapping PK off-targets, as well as empirical determination of effective concentrations by sensitive TOR kinase assays, promise to further uncover diverse biological functions of plant TOR from embryogenesis to senescence (Figure 2) [16**,20,25,26*,28,29*].
Figure 2.
The multifaceted functions of TOR kinase are uncovered by integrated chemical, genetic, genomic and metabolomics analyses. The complex TOR signaling network contributes to the regulation of plant life from embryogenesis to senescence by integrating central and secondary carbon metabolism with bioenergetics, biosynthesis, signaling, chromatin modulators, transporters, autophagy and cell cycle regulation. RNAi, RNA interference; amiRNA, artificial microRNA; RP, ribosome protein; uORF, upstream open reading frame; HSP, heat shock protein; TCA, tricarboxylic acid cycle; ETC, mitochondria electron transport chain; AGP, arabinogalactan protein; TPS, trehalose-6-phosphate synthase; TAG, triacylglycerol; XTH, xyloglucan endotransglucosylase; CYP, cytochrome P450. Torin1,2, WYE354, KU63794, PP242 and AZD8055 are specific ATP-competitive chemical inhibitors of TOR kinase.
Chemically induced TOR silencing in transgenic plants based on ethanol or estradiol induction of RNA interference (RNAi) or artificial microRNA (amiRNA) have also proven to be valuable to genetically define plant TOR functions (Figures 1–3). However, longer chemical treatment of 1–6 days and variable silencing efficacy might have resulted in more complex plant phenotypes and seemingly conflicting changes in gene expression and metabolism, which could confound interpretations of direct or indirect physiological functions of TOR signaling [6*,13**,14,15,16**, 25,31,32]. It may be possible to combine specific chemical inhibitors and silencing inducers to probe the relationship of rapid, dynamic and long-term consequences of TOR inactivation to uncover new biological insights.
TOR regulators and effectors
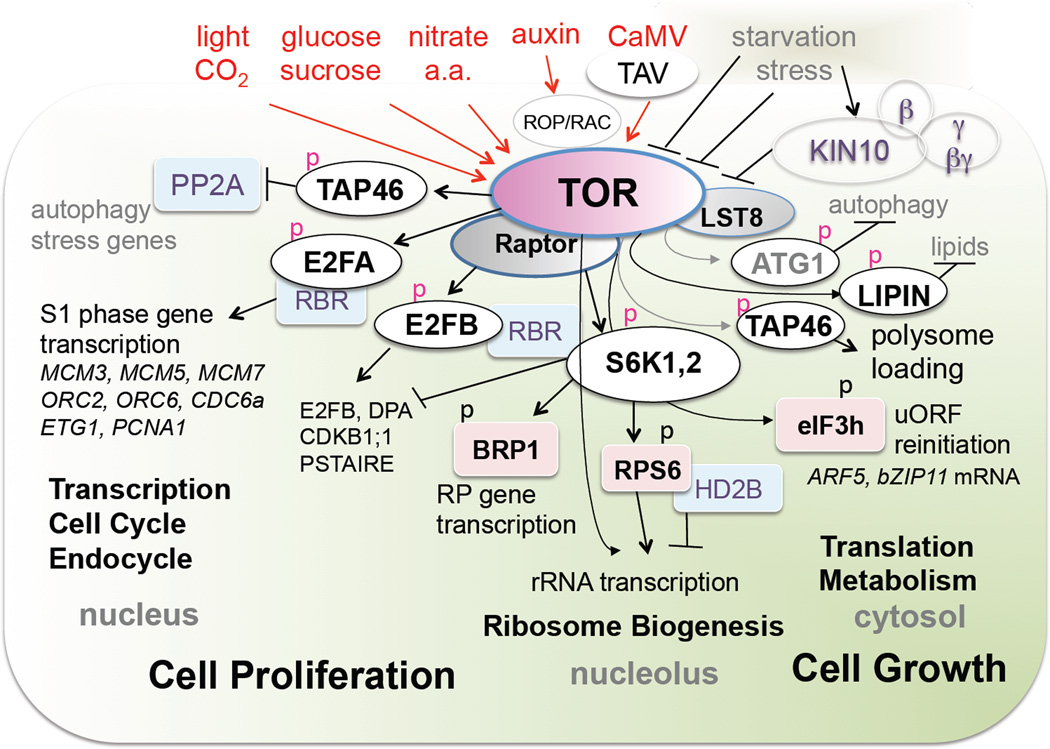
How TOR kinase senses diverse upstream regulatory signals and controls a myriad of direct or indirect downstream effectors to modulate cellular, metabolic and physiological processes are the most fascinating questions in understanding the plant TOR signaling network (Figure 3). Glucose and sucrose derived from photosynthesis stimulated by light and CO2 appear to be the most effective nutrient signals to activate plant TOR kinase [15,16**]. Glucose metabolism through glycolysis and mitochondrial electron transport chain (ETC) is essential, as glycolysis inhibitor 2-deoxyglucose and various ETC uncouplers effectively block TOR kinase activation by glucose and sucrose [16**]. Mitochondrial association, dynamic ATP elevation, and regulation by the TTT-RUVBL complex may participate in glucose-mediated TOR kinase activation [33–35]. Interestingly, nitrate and amino acids also activate TOR kinase based on S6K1 T449 phosphorylation by unknown mechanisms in Arabidopsis seedlings (Liu and Xiong, unpublished]. In addition,, auxin and viral infections have also been shown to activate TOR-S6K signaling [9**,10]. Although plants lack orthologous small guanosine 5’-triphosphatase (GTPases), Ras homolog enriched in brain (RHEB) or Rag guanosine 5’-triphosphatases (RAGs), the key upstream activators of mTORC1 [5], ROP/RAC small GTPases activated in auxin signaling could be potential upstream regulators of plant TOR kinase [36,37](Figure 3).
Figure 3.
The plant TOR signaling network. Arabidopsis TOR kinase is modulate by diverse upstream inputs and regulatory partners (Raptor and LST8) to phosphorylate S6K1,2, TAP46, E2FA, E2FB, LIPIN and ATG1 in the nucleus, nucleolus and cytosol to control transcription, cell cycle, endocycle, rRNA transcription, ribosome biogenesis, translation and metabolism, all pivotal to cell proliferation and growth. S6K, small ribosome protein 6 (RPS6) kinase; TAP46, a regulatory subunit of PP2A; E2FA/B, transcription factors; RBR, retinoblastoma repressor; HD2B, histone deacetylase2B; eIF3h, eukaryotic translation initiation factor 3h; ATG1, autophagy related kinase1.
The TORC1 acts as high molecular complexes composed of the large TOR kinase with two regulatory partners, regulatory associate protein of target of rapamycin (RAPTOR) and Lethal with Sec Thirteen protein8 (LST8) [3,5]. The growth defects of Arabidopsis raptor and lst8-1 mutants partially resemble those of various tor mutants in embryogenesis and postembryogenic growth, supporting the evolutionarily conserved function of RAPTOR and LST8 in plant TOR signaling [4,6*,14] (Figure 3). LST8 may not be required for all TOR signaling functions as the lst8-1 null mutant exhibiting more significant phenotypes under long day conditions [14]. Recent mass spectrometry analyses of mammalian RAPTOR binding partners have identified dozens of candidates besides TOR. Future studies will determine which and how these newly identified RAPTOR interacting proteins are involved in the TOR signaling network [38*].
RAPTOR interacts with the HEAT domain of TOR and regulates the activity of S6K, which serves remarkably versatile roles in multiple subcellular locals in TOR signaling [9**,24,39–41]. S6K phosphorylates BRP1 in the nucleus to activate ribosome protein gene transcription, phosphorylates RPS6 (small ribosome protein S6) in the nucleolus to enhance rRNA transcription, and acts in the cytosol to promote eukaryotic translation initiation 3h (eIF3h)-mediated translational reinitiation [9**,41,42*]. Furthermore, S6K1 also binds to retinoblastoma repressor protein (RBR) to modulate its nuclear localization, which inhibits cell cycle gene expression but promotes cell size expansion and growth [39–41] (Figure 3). Whether all of these S6K functions depend on TOR signaling requires further clarification, since other signaling pathways, such as osmotic and salt stress, and 3-phosphoinositide-dependent protein kinase1, also regulate S6K [24].
TOR kinase also directly phosphorylates TAP46 to inhibit PP2A activity, which controls stress genes, autophagy, nitrogen metabolism and protein translation [43]. Overexpression of TAP46 increases TOR activation and plant growth mainly by increasing cell size in leaves and seeds. Interestingly, TOR signaling also increases the TAP46 protein level and S6K phosphorylation through direct TAP46-S6K interaction. However, TAP46 may regulate other protein phosphatases not involved in TOR signaling and PP2A has multiple cellular functions [32]. Future studies will clarify how TOR regulates TAP46 and which PP2A functions are modulated in TOR signaling.
Since many TOR signaling functions are highly conserved in multicellular plants and animals, it has been an effective strategy to predict TOR effectors based on sequence homology and functional conservation between the plant and mammalian orthologous genes [5,6*]. It is likely that plant TOR kinase also directly phosphorylates ATG1 to inhibit autophagy [44–46], LIPIN to activate lipid synthesis [47] (Shi and Sheen, unpublished), and LARP1 (a La-domain RNA-binding protein) to promote the translation of mRNAs containing 5’-termininal oligopyrimydine (TOP) motifs (Figure 3) [38*]. Uncovering the biological relevance and functions of these conserved TOR signaling effectors will require molecular, genetic and genomic integration into the specific and unknown downstream regulatory pathways that may share little sequence or functional conservation in the photosynthetic plant system.
Although phosphoproteomic analyses by mass spectrometry has facilitated the identification of many new mTOR phosphorylation targets and signaling effectors [48,49], it remained challenging to discover transcription factors (TFs) that are the direct TOR phosphorylation targets. Based on transcriptomic analyses of the primary target genes in Arabidopsis glucose-TOR signaling, E2FA was predicted and validated to be a previously unrecognized substrate of TOR kinase [16**]. This powerful approach using systems based prediction and comprehensive experimental validation will likely play a more important role in future dissection of the TOR signaling networks in plants and animals.
Ribosome biogenesis and translational control
The most conserved functions of TOR signaling are to promote ribosome biogenesis and translation in yeast, plants and mammals in response to nutrients and growth regulators [3–5, 6*]. Consistently, glucose-TOR signaling activates more than 100 primary target genes encoding ribosomal proteins (RPS and RPL), ribosomal RNA processing proteins, ribosome biogenesis regulatory protein, and protein initiation and elongation factors in Arabidopsis [14,16**]. Interestingly, these genes are also primary target genes of Arabidopsis energy sensor kinase KIN10 repressed by glucose but activated by starvation and stress [50], suggesting their intimate antagonistic regulation (Figure 3). The HEAT domain of TOR directly binds to 45S rRNA gene promoter and 5’ external transcribed spacer elements to promote rRNA synthesis and ribosome biogenesis stimulated by light, sugar and nitrogen nutrients [15,20]. A new study using MALTI-TOF mass spectrometry identified a plant-specific histone deacetylase2B (HD2B) that directly interacts with RPS6 in Arabidopsis. Analyses in protoplast assays and in transgenic rps6b mutant plants suggest that HD2B and RPS6 form complexes to negatively regulate rRNA transcription in the nucleolus. RPS6 phosphorylation by TOR-S6K signaling likely relieves the repression. Interestingly, chromatin immunoprecipitation (ChIP) PCR analyses indicated that TOR and RPS6 bind to distinct sites on the rRNA gene promoter to mediate transcriptional and epigenetic control, respectively [20,41,42*]. TOR-S6K signaling may also activate BRP1 TF and RP gene expression to promote ribosome biogenesis in response to nutrient availability and auxin stimulation for cell expansion [41](Figure 3).
Plant translational control involves both conserved and plant-specific regulation [51]. TOR-TAP46 signaling is indispensible for global translation in plant cells by unknown molecular mechanisms that require future characterization [43]. It will also be of great interest to explore whether the recently discovered role of TOR-LARP1 signaling in mediating TOP mRNA translation in mammals has any parallel in plants [38*,51,52]. However, light appears to enhance the translation of plant mRNAs with a cis-element in their 5’ UTR distinct from TOP motifs [53]. Future research may connect TOR signaling to translational control during photomorphogenesis unique in plants.
Although mTOR-S6K signaling plays multiple roles in translational control [27], how plant S6K regulates translation in the cytosol remains unclear. Recent efforts have discovered novel TOR-S6K signaling functions in translational reinitiation of specific viral and plant mRNAs. The Cauliflower Mosaic Virus (CaMV) mRNA translational reinitiation is enhanced by a virus protein TAV (translational transactivator/viroplasmin). TAV stimulates TOR hyperactivation and S6K1 phosphorylation to promote TOR association of polysomes with eIF3 complex, and RISP (reinitiation supporting protein) phosphorylation and activation. Consistently, TOR-deficient plants are resistant to viral infection [10]. However, the requirement of TOR signaling for successful infection is not universal by all plant pathogens. For example, for different plant potyviruses, infection by Watermelon Mosaic Virus is prevented by TOR inhibitor AZD8055 and TOR RNAi but not for Turnip Mosaic Virus [54]. Different plant pathogens likely have evolved multiple mechanisms to gain access to nutrient and energy resources in plants.
Auxin was also shown recently to promote polysome loading of mRNAs containing upstream open reading frames (uORF). The mRNAs encoding ARF5 or bZIP11 contain uORF and are stimulated by auxin for efficient translational reinitiation via TOR-S6K activation and eIF3h phosphorylation [9**]. As S6K1 is dissociated from but TOR is recruited to the polysome-eIF3h complex after auxin treatment, it remains to be demonstrated that S6K1 but not TOR kinase directly phosphorylates and activates eIF3h at S178. Since auxin regulation of primary gene transcription is not affected in null tor mutant seedlings, auxin promotion of translational reinitiation of ARF activators and repressors may be associated with specific but not all auxin responses [9**,16**]. It will also be interesting to determine how auxin activates whereas sucrose represses bZIP11 mRNA translational reinitiation through uORF2 in the 5’UTR, even though both auxin and sucrose activate TOR-S6K signaling in plants (Figure 3) [9**,16**,55].
Cell cycle and cell size regulation
Rapamycin arrested yeast and mammalian cells in the G1 phase of cell cycle. Arabidopsis null tor mutants were arrested during early embryogenesis and prevented postembryonic seedling development. The precise molecular mechanisms controlling the cell cycle by TOR signaling remained mostly unknown for decades [3–5,15,25]. Genome-wide analyses of early glucose-TOR signaling target genes in WT and inducible null tor plants have provided clear evidence that Arabidopsis TORC1 is directly involved in a broad spectrum of cell cycle gene activation [13**,16**]. Systems, cellular, biochemical and genetic analyses revealed that glucose-TOR signaling phosphorylates and activates E2FA TF to enhance S phase gene transcription and DNA replication in the primary root meristem independent of auxin and cytokinin signaling and the conventional CYCLIND (CYCD)-CYCLIN-DEPENDENT KINASE (CDK)-RBR pathway [56]. The e2fa mutant showed diminished glucose activation of EdU (5-ethynyk-2’-deoxyuridine) labeling in the root stem cells, meristem and elongation zone, suggesting dual functions in cell cycle and endocycle regulation. This new mechanism of cell cycle activation by TOR kinase to mediate nutrient signaling is critical at the developmental transition from lipid-based heterotrophic to sugar-based photoautotrophic growth vital to postembryonic plant growth [16**]. As TOR kinase also phosphorylates and activates E2FB and other TFs to promote cell cycle, TOR signaling is not restricted to the primary root meristem and may play key roles in the cell cycle regulation in the shoot apical meristem and other plant tissues and organs [57]. Besides direct TOR phosphorylation, glucose-TOR signaling also activates genes encoding root growth peptides and S-assimilation pathway required for promoting cell cycle, but represses UPBEAT TF, a negative regulator of cellular proliferation via redox processes [16**,58].
Interestingly, plant TOR signaling may activate S6K to inhibit cell cycle via promoting RBR nuclear localization to interact and repress E2FB. In Arabidopsis suspension cells expressing S6K1-RNAi, E2FB, DPA, CDKB1;1 and PSTAIRE proteins are significantly elevated, likely through mutually antagonistic S6K-E2FB protein stability control and escape from transcriptional repression by RBR-E2FB. S6K’s role in glucose-TOR signaling appears to predominantly promote cell growth but inhibit cell cycle. However, the later may change in different cellular contexts, as S6K is also involved in multiple functions with different partners in three subcellular compartments for transcriptional and translation controls (Figure 3) [17,27,39–41]. As RPS6 is a key substrate of S6K, Arabidopsis rps6a and rps6b mutant plants have reduced leaf, root and inflorescence size and epidermal cell size, but are more resistant to rapamycin inhibition of growth in FKPB12 overexpression plants [15, 42*]. Interestingly, TOR overexpression and FKBP overexpression plants treated with rapamycin exhibit accelerated and reduced cell death and senescence in an RPS6 dependent manner, respectively [15]. How TOR-S6K-RPS6 signaling regulates cellular growth and senescence in different plant organs will be an important future research direction.
Future challenges
The biological functions of plant TOR in embryogenesis, seedling and plant growth, metabolism, and senescence have emerged. The molecular regulatory mechanisms of the plant TOR signaling network are starting to be elucidated in the root meristems and growing and differentiated cells. The application of versatile chemical tools and integrated systems, cellular, genetic, genomic and phosphoproteomic analyses will facilitate the discoveries of new regulators and molecular links in TOR signaling. Major puzzles waiting to be resolved include how TOR kinase modulates an increasingly large array of downstream effectors in response to distinct upstream signals and regulators (Figure 3). Nutrient and energy signaling functions are fundamental to transcriptional, translational and metabolic controls in all living cells. It will be important to expand our knowledge on how ubiquitously expressed TOR are regulated by diverse input signals and how TOR activation and repression are coordinated in different cellular contexts that are proliferating, expanding, differentiating or fully differentiated in plant’s daily life. Development of new and sensitive technologies for single-cell based genetic and chemical perturbation and for gene expression and metabolite profiling will be desired.
As TOR kinase is an integral part of the glucose signaling network in photosynthetic plants [8], it is important to understanding how the dynamic processes and regulation in sugar production, transport, storage and metabolism [59,60] modulate TOR signaling in different plant cells, tissues and organs, which form the basis of plant growth and developmental programs. Although transcriptomic and metabolomic studies have implicated a major role of TOR signaling in regulating primary and secondary plant metabolism [13**,14,15,16**,31], a gap exists between the regulation of TOR target genes and enzymes and the long-term steady-state metabolite accumulation. Dynamic profiling of plant metabolite changes together with flux and enzymatic measurement [61] after short term TOR inactivation may offer new insights. Finally, much information will be gained in understanding the plant energy and stress signaling network by elucidating the antagonistic functions of TOR and KIN10 as key energy sensors and central regulators of transcriptional, translational and metabolic programs in response to nutrients, hormones and environmental cues.
Highlights.
TOR integrates nutrient and energy signaling to promote cell division and growth.
Powerful chemical tools are developed for probing plant TOR functions.
Both conserved and unique TOR effectors are identified in the plant system.
Acknowledgements
We apologize for limited literature coverage due to space limitation. The projects on the plant TOR signaling network have been supported by the NIH grants and WJC Special Project RDA-Korea to J.S. Y.X. is supported by Chinese Academy of Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Aramburu J, Ortells MC, Tejedor S, Buxade M, Lopez-Rodriguez C. Transcriptional regulation of the stress response by mTOR. Sci Signal. 2014;7:re2. doi: 10.1126/scisignal.2005326. [DOI] [PubMed] [Google Scholar]
- 2.Hardie DG. AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol. 2014;33:1–7. doi: 10.1016/j.ceb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robaglia C, Thomas M, Meyer C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol. 2012;15:301–307. doi: 10.1016/j.pbi.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–162. doi: 10.1038/nrm3757. [DOI] [PubMed] [Google Scholar]
- 6. Xiong Y, Sheen J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014;164:499–512. doi: 10.1104/pp.113.229948. This comprehensive review summarized the multifaceted roles of TOR signaling in plant growth and development, and the molecular mechanisms underlying TOR-mediated ribosomal biogenesis, translation, metabolism and transcription.
- 7.Perez-Perez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010;152:1874–1888. doi: 10.1104/pp.109.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sheen J. Master Regulators in Plant Glucose Signaling Networks. 2014;57:67–79. doi: 10.1007/s12374-014-0902-7. This review specially focuses on three glucose master regulators: HXK1, KIN10/11 and TOR for their integrative and complementary roles from cellular signaling and metabolism to development in plant glucose signaling networks.
- 9. Schepetilnikov M, Dimitrova M, Mancera-Martinez E, Geldreich A, Keller M, Ryabova LA. TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J. 2013;32:1087–1102. doi: 10.1038/emboj.2013.61. This study reveals that auxin can enhance TOR activity to promote the translation reinitiation of uORF containing mRNA via S6K1 phosphorylation of eIF3h. Accordingly, TOR signaling deficiency impaired auxin mediated root gravitropism.
- 10.Schepetilnikov M, Kobayashi K, Geldreich A, Caranta C, Robaglia C, Keller M, Ryabova LA. Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO J. 2011;30:1343–1356. doi: 10.1038/emboj.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira AP, Ludwig C, Zampieri M, Weisser H, Aebersold R, Sauer U. Dynamic phosphoproteomics reveals TORC1-dependent regulation of yeast nucleotide and amino acid biosynthesis. Sci Signal. 2015;8:rs4. doi: 10.1126/scisignal.2005768. [DOI] [PubMed] [Google Scholar]
- 13. Caldana C, Li Y, Leisse A, Zhang Y, Bartholomaeus L, Fernie AR, Willmitzer L, Giavalisco P. Systemic analysis of inducible target of rapamycin mutants reveal a general metabolic switch controlling growth in Arabidopsis thaliana. Plant J. 2013;73:897–909. doi: 10.1111/tpj.12080. Using the estradiol-inducible amiR-tor mutants, the authors studied the effect of long term and partial disruption of TOR signaling by global transcriptomic and metabolite profiling, revealing a key role of TOR in primary and secondary metabolism.
- 14.Moreau M, Azzopardi M, Clement G, Dobrenel T, Marchive C, Renne C, Martin-Magniette ML, Taconnat L, Renou JP, Robaglia C, et al. Mutations in the Arabidopsis homolog of LST8/GbetaL, a partner of the target of Rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell. 2012;24:463–481. doi: 10.1105/tpc.111.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren M, Venglat P, Qiu S, Feng L, Cao Y, Wang E, Xiang D, Wang J, Alexander D, Chalivendra S, et al. Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell. 2012;24:4850–4874. doi: 10.1105/tpc.112.107144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature. 2013;496:181–186. doi: 10.1038/nature12030. The authors made a surprising discovery that TOR can sense and transduce shoot photosynthesis-derived glucose signal through glycolysis and mitochondrial energy relay, to rapidly control global transcriptome reprogramming for root meristem activation. Unexpectedly, TOR directly phosphorylates and activates E2Fa to induce S-phase genes. This represented an unconventional mode of TOR beyond stimulating the translation of proteins involved in cell cycle progression through S6K1 and 4E-BP in mammals.
- 17.Henriques R, Bogre L, Horvath B, Magyar Z. Balancing act: matching growth with environment by the TOR signalling pathway. J Exp Bot. 2014;65:2691–2701. doi: 10.1093/jxb/eru049. [DOI] [PubMed] [Google Scholar]
- 18.Dobrenel T, Marchive C, Azzopardi M, Clement G, Moreau M, Sormani R, Robaglia C, Meyer C. Sugar metabolism and the plant target of rapamycin kinase: a sweet operaTOR? Front Plant Sci. 2013;4:93. doi: 10.3389/fpls.2013.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci U S A. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren M, Qiu S, Venglat P, Xiang D, Feng L, Selvaraj G, Datla R. Target of rapamycin regulates development and ribosomal RNA expression through kinase domain in Arabidopsis. Plant Physiol. 2011;155:1367–1382. doi: 10.1104/pp.110.169045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crespo JL, Diaz-Troya S, Florencio FJ. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005;139:1736–1749. doi: 10.1104/pp.105.070847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee do Y, Fiehn O. Metabolomic response of Chlamydomonas reinhardtii to the inhibition of target of rapamycin (TOR) by rapamycin. J Microbiol Biotechnol. 2013;23:923–931. doi: 10.4014/jmb.1304.04057. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Perez ME, Florencio FJ, Crespo JL. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010;152:1874–1888. doi: 10.1104/pp.109.152520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahfouz MM, Kim S, Delauney AJ, Verma DP. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong Y, Sheen J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J Biol Chem. 2012;287:2836–2842. doi: 10.1074/jbc.M111.300749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341:1236566. doi: 10.1126/science.1236566. This work nicely demonstrated that the intrinsic capacity of a phosphorylation site is a major determinant of mTOR substrates’ sensitivity to rapamycin and the modulators of the pathway. Modification of the phosphorylation site is sufficient to alter its rapamycin sensitivity.
- 27.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nature Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 28.Liu Q, Kirubakaran S, Hur W, Niepel M, Westover K, Thoreen CC, Wang J, Ni J, Patricelli MP, Vogel K, et al. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J Biol Chem. 2012;287:9742–9752. doi: 10.1074/jbc.M111.304485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montane MH, Menand B. ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot. 2013;64:4361–4374. doi: 10.1093/jxb/ert242. This work analyzed the effect of ATP-competitive mTOR inhibitors in plant growth and development, and found that these inhibitors cause growth defects resembling tor mutants in a TOR gene dosage dependent manner.
- 30.Wang D, Harper JF, Gribskov M. Systematic Trans-Genomic Comparison of Protein Kinases between Arabidopsis and Saccharomyces cerevisiae. Plant Physiol. 2003;132:2152–2165. doi: 10.1104/pp.103.021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolai M, Bedu M, Robaglia C, Meyer C. The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 2007;8:864–870. doi: 10.1038/sj.embor.7401043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn CS, Ahn HK, Pai HS. Overexpression of the PP2A regulatory subunit Tap46 leads to enhanced plant growth through stimulation of the TOR signalling pathway. J Exp Bot. 2014;66:827–840. doi: 10.1093/jxb/eru438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 34.Kim SG, Hoffman GR, Poulogiannis G, Buel GR, Jang YJ, Lee KW, Kim BY, Erikson RL, Cantley LC, Choo AY, et al. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol Cell. 2013;49:172–185. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramanathan A, Schreiber SL. Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci U S A. 2009;106:22229–22232. doi: 10.1073/pnas.0912074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao LZ, Cheung AY, Wu HM. Plant Rac-like GTPases are activated by auxin and mediate auxin-responsive gene expression. Plant Cell. 2002;14:2745–2760. doi: 10.1105/tpc.006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusova H, et al. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science. 343:1025–1028. doi: 10.1126/science.1245125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang HD, Jensen JM, Diao IT, et al. La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1) J Biol Chem. 2015 doi: 10.1074/jbc.M114.621730. This work identified LARP1 as a direct TORC1 substrate, which can directly bind 5’TOP motif of mRNA to repress their translation. TOR phosphorylation of LARP1 releases this repression to enhance the mRNA translation.
- 39.Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bogre L. Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J. 2010;29:2979–2993. doi: 10.1038/emboj.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henriques R, Magyar Z, Bogre L. S6K1 and E2FB are in mutually antagonistic regulatory links controlling cell growth and proliferation in Arabidopsis. Plant Signal Behav. 2013;8:e24367. doi: 10.4161/psb.24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin YJ, Kim S, Du H, Choi S, Verma DP, Cheon CI. Possible dual regulatory circuits involving AtS6K1 in the regulation of plant cell cycle and growth. Mol Cells. 2012;33:487–496. doi: 10.1007/s10059-012-2275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim YK, Kim S, Shin YJ, Hur YS, Kim WY, Lee MS, Cheon CI, Verma DP. Ribosomal protein S6, a target of rapamycin, is involved in the regulation of rRNA genes by possible epigenetic changes in Arabidopsis. J Biol Chem. 2014;289:3901–3912. doi: 10.1074/jbc.M113.515015. The authors uncovered that Arabidopsis RPS6 interacts with histone deacetylase 2B (AtHD2B), and binds directly to the promoter of rRNA gene, which indicates an important role of TOR signaling in epigenetic regulation of rDNA transcription and ribosome biogenesis in plants.
- 43.Ahn CS, Han JA, Lee HS, Lee S, Pai HS. The PP2A regulatory subunit Tap46, a component of the TOR signaling pathway, modulates growth and metabolism in plants. Plant Cell. 2011;23:185–209. doi: 10.1105/tpc.110.074005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li F, Vierstra RD. Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012;17:526–537. doi: 10.1016/j.tplants.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Bassham DC. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. doi: 10.1146/annurev-arplant-042811-105441. [DOI] [PubMed] [Google Scholar]
- 47.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- 51.Browning KS, Bailey-Serres J. Mechanism of cytoplasmic mRNA translation. Arabidopsis Book. 2015;13:e0176. doi: 10.1199/tab.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu MJ, Wu SH, Chen HM. Widespread translational control contributes to the regulation of Arabidopsis photomorphogenesis. Mol Syst Biol. 2012;8:566. doi: 10.1038/msb.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ouibrahim L, Giner Rubio A, Moretti A, Montane MH, Menand B, Meyer C, Robaglia C, Caranta C. Potyviruses differ in their requirement for TOR signaling. J Gen Virol. 2015 doi: 10.1099/vir.0.000186. [DOI] [PubMed] [Google Scholar]
- 55.Lastdrager J, Hanson J, Smeekens S. Sugar signals and the control of plant growth and development. J Exp Bot. 2014;65:799–807. doi: 10.1093/jxb/ert474. [DOI] [PubMed] [Google Scholar]
- 56.Magyar Z, Horvath B, Khan S, Mohammed B, Henriques R, De Veylder L, Bako L, Scheres B, Bogre L. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012;31:1480–1493. doi: 10.1038/emboj.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiong Y, Sheen J. Moving beyond translation: glucose-TOR signaling in the transcriptional control of cell cycle. Cell Cycle. 2013;12:1989–1990. doi: 10.4161/cc.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 59.Ruan YL. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu Rev Plant Biol. 2014;65:33–67. doi: 10.1146/annurev-arplant-050213-040251. [DOI] [PubMed] [Google Scholar]
- 60.Stitt M. Progress in understanding and engineering primary plant metabolism. Curr Opin Biotechnol. 2013;24:229–238. doi: 10.1016/j.copbio.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Stitt M, Sulpice R, Keurentjes J. Metabolic networks: how to identify key components in the regulation of metabolism and growth. Plant Physiol. 2010;152:428–444. doi: 10.1104/pp.109.150821. [DOI] [PMC free article] [PubMed] [Google Scholar]