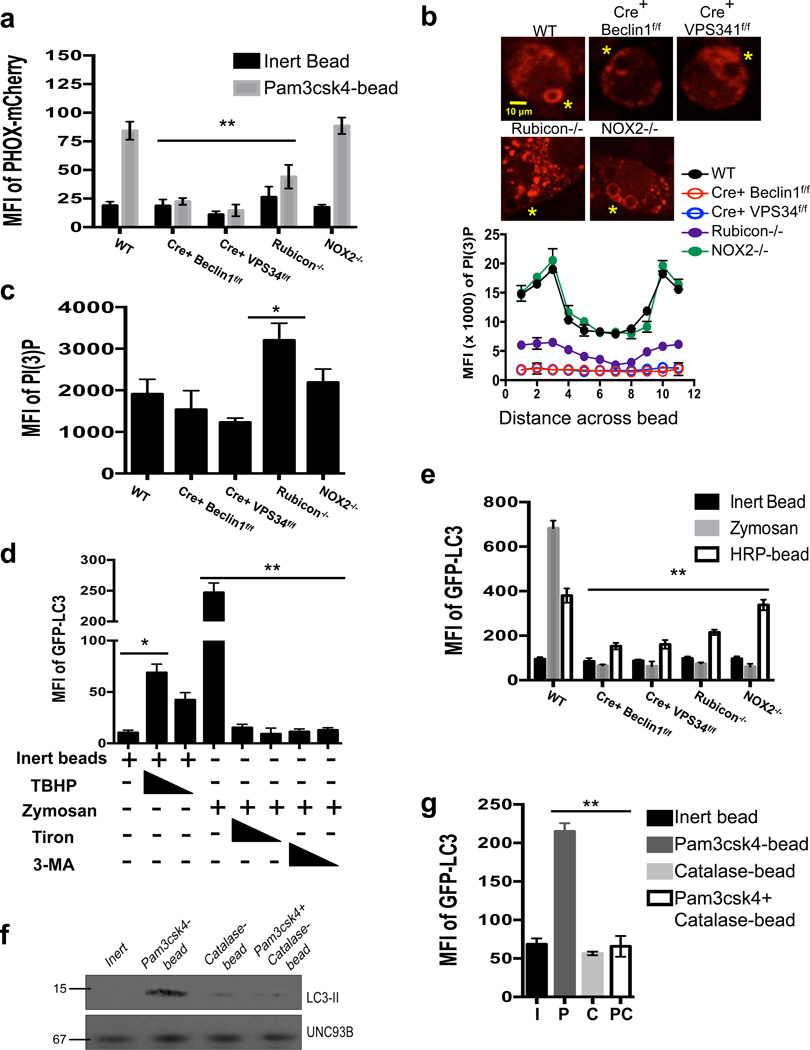
Figure 4. PI(3)P and ROS are both required for LAP.
(a–c) Bone marrow-derived macrophages from genetic knockout strains were transiently transfected with PX-mCherry. After 48 hours of transfection, cells were fed inert beads or Pam3csk4-beads for 30 minutes. PI(3)P on LAPosomes was analyzed by flow cytometry (a) or confocal microscopy (b–c). Yellow asterisks indicate internalized beads. Representative images and signal intensity profiles for PX-mCherry across phagocytosed beads are quantified and shown graphically (n ≥ 25 / genotype) (b). Masks were generated around whole cells and the MFI of PI(3)P within the cell was quantified using Slidebook software. Data are presented as mean ± SD of three independent experiments (**p < 0.001) (c). Data are presented as mean ± SD of two independent experiments (n ≥ 25 / genotype). (d) RAW-GFP-LC3 cells were fed inert beads or zymosan, in the presence or absence of tert-butyl hydroperoxide (TBHP, 100 µM, 50 µM), Tiron (1 mM, 0.5 mM), or 3-MA (25 mM, 5 mM). Translocation of GFP-LC3 to the LAPosome was assessed at 1 h by flow cytometry. (e) Bone marrow-derived macrophages from GFP-LC3+ genetic knockout strains were fed inert beads, Alexa Fluor 594-zymosan, or HRP-coupled beads. Translocation of GFP-LC3 to the LAPosome was assessed at 1 h by flow cytometry. Data are presented as mean ± SD of three independent experiments (**p < 0.001) (f–g) RAW-GFP-LC3 cells were fed inert beads (I), Pam3csk4-beads (P), Catalase-beads (C), or Pam3csk4+Catalase-beads (PC). Translocation of GFP-LC3 to the LAPosome was assessed at 1 h by immunoblot analysis of purified phagosomal proteins (f) and flow cytometry (g). Data are presented as mean ± SD of three independent experiments (**p < 0.001).