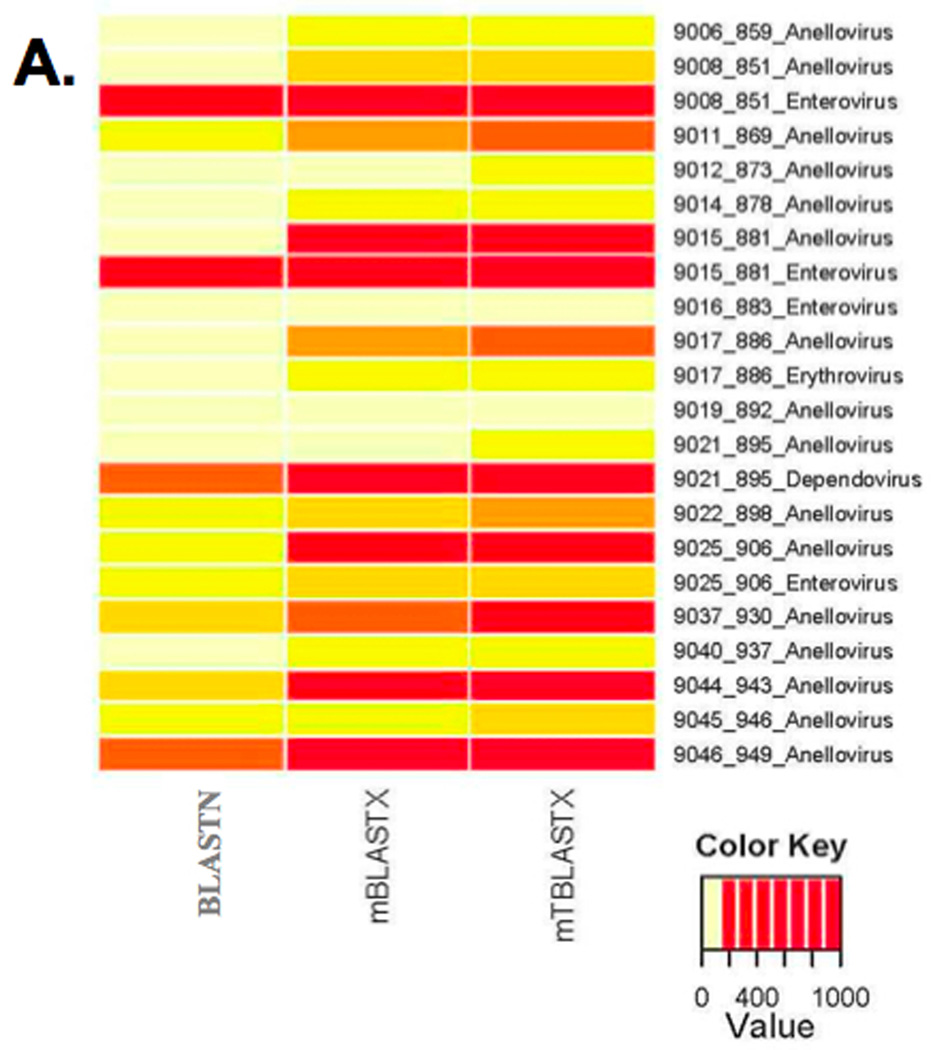
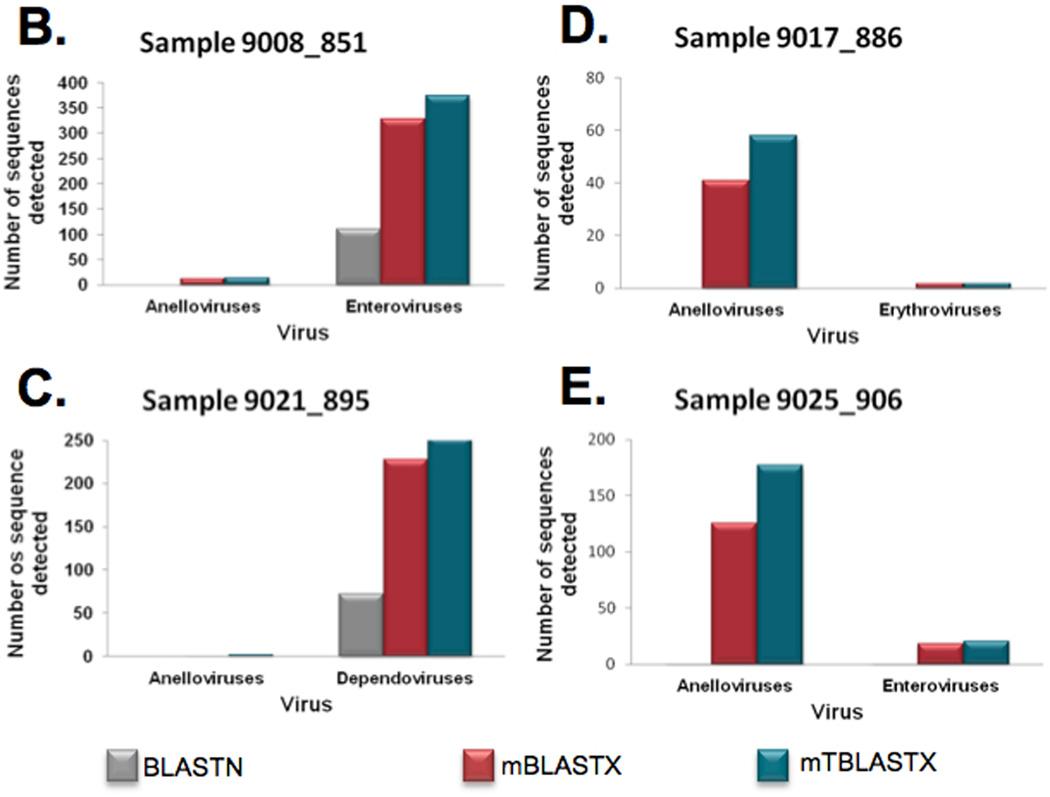
Figure 7.
Metagenomic shotgun reads from 20 blood plasma samples from febrile children were aligned to viral reference genomes at the nucleotide sequence level and compared to translated alignments generated using mBLASTX and mTBLASTX. (A) Translated alignments improve detection of ssDNA and ssRNA viruses in metagenomic samples. (B–E) Examples from 4 plasma samples from febrile children and the detected Anelloviruses (ssDNA genomes), enteroviruses (+ssRNA genomes), erythroviruses (ssDNA genomes), and dependoviruses (ssDNA genomes). The numbers of viral sequences detected by each alignment method are shown.