Figure 2.
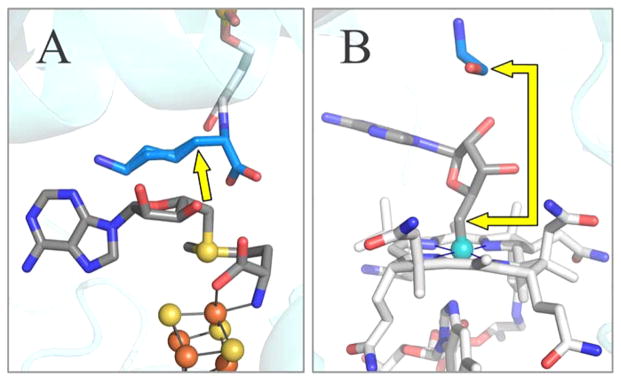
Active site structure of LAM (A) and ethanolamine ammonia-lyase (EAL) (B). The illustration of EAL contains the B12 cofactor as modeled from the crystal structure of EAL with the analogue adeninylpentylcobalamin. The yellow arrows show the distance from C5′ prior to S–C5′ or Co–C5′ bond cleavage, to target carbon atom in the substrate. PDB IDs are 2A5H (LAM) and 3ABS (ethanolamine ammonia-lyase). Carbon atoms are colored as: blue for substrate, dark grey for adenosine and SAM, and light grey for cobalamin and PLP.
