Abstract
Mitochondrial DNA (mtDNA) is widely used to clarify phylogenetic relationships among and within species, and to determine population structure. Due to the linked nature of mtDNA genes it is expected that different genes will show similar results. Phylogenetic incongruence using mtDNA genes may result from processes such as heteroplasmy, nuclear integration of mitochondrial genes, polymerase errors, contamination, and recombination. In this study we used sequences from two mitochondrial genes (cytochrome b and cytochrome oxidase subunit I) from the wild vectors of Chagas disease, Triatoma eratyrusiformis and Mepraia species to test for topological congruence. The results showed some cases of phylogenetic incongruence due to misplacement of four haplotypes of four individuals. We discuss the possible causes of such incongruence and suggest that the explanation is an intra-individual variation likely due to heteroplasmy. This phenomenon is an independent evidence of common ancestry between these taxa.
Keywords: heteroplasmy, sylvatic vectors, Chagas disease, gajardoi, spinolai, cytochrome
Introduction
The simplicity of maternal inheritance without bi-parental recombination, high rates of variability and large copy numbers are key features that make mitochondrial DNA (mtDNA) highly useful in resolving questions related to phylogenetic and phylogeographic relationships (Avise et al., 1987; Zhang et al., 1995). However, certain processes such as heteroplasmy, nuclear integration of mitochondrial genes, polymerase errors, DNA contamination, ancestral polymorphisms and recombination may lead to incongruence using independent loci. For example, heteroplasmy, the presence of more than one haplotype within a single organism (Boyce et al., 1989; Frey and Frey, 2004) may result in incongruence of phylogenetic topologies and hence negatively impact the resolution of evolutionary relationships of organisms. Heteroplasmy has been reported in insects and other arthropods (Frey and Frey, 2004; Fontaine et al., 2007; Magnacca and Brown, 2010; Nunes et al., 2013). Another source of potential phylogenetic ambiguity is the nuclear integration of mitochondrial gene fragments, which has been suggested to occur in Triatominae (Dotson and Beard, 2001) and other arthropods (Zhang and Hewitt, 1996a; Parfait et al., 1998; Bensasson et al., 2001).
Triatomines of the genus Mepraia (Mazza et al., 1940) are blood-sucking insects that play an important role in the transmission of Trypanosoma cruzi, the etiologic agent of Chagas disease in the sylvatic cycle (Rozas et al., 2007; Botto-Mahan et al., 2008). Mepraia is endemic to semiarid and arid regions, and is distributed in coastal and interior valleys of northern and central Chile (Frías et al., 1998; Campos et al., 2013a). Their distribution in wild and peridomestic habitats, their opportunistic feeding behavior and human settlement in risk areas are features of high epidemiological significance as potential vectors for humans (Cattan et al., 2002; Toledo et al., 2013). Three species are currently included in the genus: M. spinolai Porter 1943, M. gajardoi and M. parapatrica (Frías et al., 1998; Frías, 2010). M. parapatrica has been recently described based on morphology (Frías, 2010). Mitochondrial gene sequences also support three lineages congruent with the three described species (Campos et al., 2013a) However, the specific status of M. parapatricaremains controversial, given the widely recognized morphological plasticity within the subfamily Triatominae (Dujardin et al., 1999) and the presumptive introgression/hybridization processes acting within Mepraia (Calleros et al., 2010; Campos et al., 2011). The genus Mepraia belongs to the spinolai complex together with Triatoma eratyrusiformis Del Ponte 1929 and Triatoma breyeri Del Ponte 1929. The last two taxa are geographically separated from Mepraia species by the Andes Range (Lent and Wygodzinsky, 1979). T. eratyrusiformis is closely related to Mepraia (Lent and Wygodzinsky, 1979; Hypa et al., 2002; Moreno et al., 2006; Frías, 2010; Campos et al., 2013b). The monophyly of the spinolai complex is supported by mitochondrial gene sequences (Campos et al., 2013b; Justi et al., 2014), and their divergence on both sides of the Andes from the common ancestor probably occurred after the uplift of the Andes during the Miocene (Moreno et al., 2006; Frías, 2010; Campos et al., 2013a,b). Although several studies have reported heteroplasmy in arthropods, this phenomenon seems to be underestimated because conventional (automated) sequencing may fail to detect it (Dos et al., 2008; Magnacca and Brown, 2010). In this study, we found incongruence in phylogenies inferred with two mitochondrial markers, cytochrome b (cyt b) and cytochrome oxidase subunit-I (COI) in Mepraiaspecies and T. eratyrusiformis and discuss the origin that may have produced this pattern.
Materials and Methods
Sample collection
We used 66 mitochondrial gene sequences of COI and cyt b of Mepraia (GenBank accession numbers KC236913-KC236978, Campos et al., 2013a). Ten additional sequences of both genes were also included: M. spinolai from Til Til, Metropolitan Region (33°06′19″ S; 70°55′53″ W, N = 2) and Los Andes, Valparaíso Region (32°01′33′ S; 70°04′16″ W; N = 2); T. eratyrusiformis from Salinas de Bustos, Departmento of Independencia, Province of La Rioja, Argentina (N = 4); Triatoma infestans (N = 2) was used as outgroup, (accession numbers KM258433-KM258442).
Insects were manually collected by trained people as follows: a person wearing safety clothes and carrying collecting tools (forceps, brush, plastic can) was used as bait, waiting 15–20 min in areas with ecological attributes to harbor kissing-bugs; if no kissing-bug appeared, the investigator moved to another sampling site. Bugs were transported to the laboratory; the limbs were dissected and kept in 70% ethanol at −20 °C.
Mitochondrial DNA extraction, amplification, and sequencing
Genomic DNA from legs was extracted using the DNA extraction kit E.Z.N.A. Tissue DNA® (Omega Biotec, Georgia) according to manufacturer’s instructions. A 636-bp fragment of the mitochondrial cytochrome oxidase subunit-I (COI) gene, and a 682-bp fragment of the cytochrome b (cyt b) gene were amplified via polymerase chain reaction (PCR) using Platinum® Taq DNA polymerase (Invitrogen, Brazil) and the primers 7432 (forward) (5′-GGACGWGGWATTTATTATGGATC-3′) and 7433 (reverse) (5′-GCWCCAATTCARGTTARTAA-3′) for cyt b (Monteiro et al., 2003) and the primers LCO1490 (forward) (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (reverse) (5′-TAAACTTCAGGGTGAC CAAAAAATCA-3′) for COI (Folmer et al., 1994). The following conditions were used to amplify both cyt b and COI: an initial denaturation at 94 °C for 3 min, 30 cycles of 1 min at 94 °C, 45 °C for 1 min and 72 °C for 1 min, followed by a final extension of 10 min. Verification of successful amplification was assessed by 2% agarose gel electrophoresis.
Sequencing reactions were conducted by Macrogen Inc. (South Korea) using the same PCR primers. Sequences were edited using Bioedit 7.0.8.0 (Hall, 1999) and aligned using Clustal W (Thompson et al., 1994) as implemented in Bioedit (Hall, 1999). After the alignments, sites that showed nucleotide substitutions were re-examined by visual inspection of each individual’s raw chromatogram. For phylogenetic reconstruction (see below) ambiguous bases were coded using the nucleotide ambiguity code (IUPAC). Non-synonymous substitutions and stop codons were checked using DnaSp 5.1 (Librado and Rozas, 2009). The resulting COI data was 508 bp in length and cyt b was 514 bp.
Phylogenetic analyses
Phylogenetic reconstructions were performed separately for each mitochondrial gene by maximum likelihood (ML) and maximum parsimony (MP). ML analyses were performed using the online platform PhyML 3.0 (Guindon et al., 2010). The best-fitting model of nucleotide substitution was selected using the Akaike information criterion (Akaike, 1974) on cyt b (TrN +I, I: 0.66) and COI (HKY+G, G: 0.068) implemented in the program jmodelTest 0.1.1 (Posada, 2008). MP analyses were performed using PAUP* 4.0b10 (Swofford, 2002) with the heuristic search option and tree bisection reconnection (TBR) branch swapping. Nodal supports were estimated by the bootstrap method (Felsenstein, 1985) with 1000 replicates. We considered branches receiving > 70% bootstrap support to be well-supported (Hillis et al., 1993; Wilcox et al., 2002). Trees were visualized using the FigTree v. 1.1.2 program. Triatoma infestans, was used as outgroup based on its phylogenetic proximity to the spinolai complex (Hypa et al., 2002; de Paula et al., 2005; Campos et al., 2013b). This research was undertaken with approval from the Bioethics Committee of the Pontificia Universidad Católica de Valparaíso, Chile.
Results
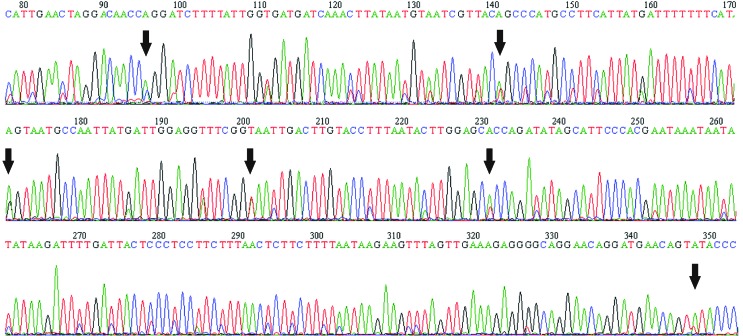
Ambiguous bases in the chromatograms were observed in the samples Tera1 and 53Til for the COI gene, and in the Tera2 sample for the cyt b gene. Samples Tera1 and Tera2 were reamplified and sequenced for both genes. The sample 53Til was not again amplified due to limited genomic DNA availability. The second round of sequencing for the cyt b gene did not show differences with the first round for the Tera2 sample, however some nucleotide differences were observed in the Tera1 sample. The second round of sequencing for the COI gene amplification did not show differences in the Tera2 sample, while six sites showed nucleotide differences in the Tera1 sample. All nucleotide differences between the first and the second round of sequencing for the Tera1 and Tera2 samples were observed in sites with ambiguous bases (Figure 1). Analyses of the cyt b and COI genes showed no stop codons. The cyt b and COI alignments showed 155 and 125 variable polymorphic sites, respectively.
Figure 1. Chromatogram of a segment of the COI gene after the second sequencing round for Tera1. Arrows indicate the sites with differences compared with the first sequencing round.
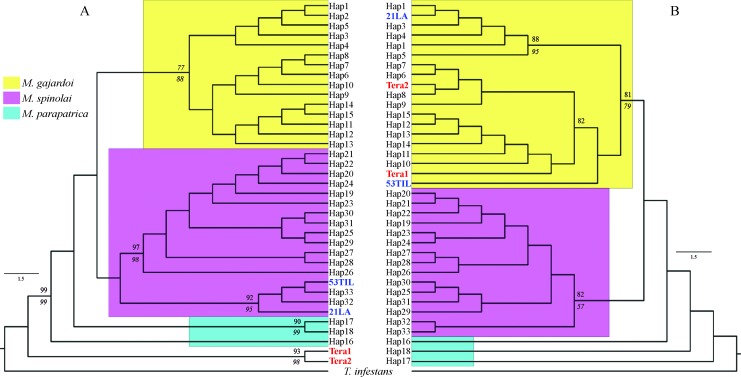
Tree topologies performed with cyt b and COI genesare shown in the Figure 2. Both phylogenetic topologies showed similar results, although incongruence was observed with four haplotypes intermingled among lineages. The cladogram reconstructed with the COI gene sequence (Figure 2B) showed two haplotypes of M. spinolai (53Til and 21LA) grouped within the M. gajardoi lineage, and two haplotypes of T. eratyrusiformis (Tera1 and Tera2) were also included within the M. gajardoi lineage (81% bootstrap support). These haplotypes were grouped as expected according to their taxonomy and geography in the cladogram reconstructed with cyt b, leading to conclude that the samples Tera1 and Tera2 represent sister species of Mepraia and the samples 53Til and 21LA belong to M. spinolai (Figure 2A). In the incongruent sequences (53Til, 21LA, Tera1, and Tera2) non-synonymous mutations were not observed in the COI gene.
Figure 2. Maximum likelihood phylogenetic trees of Mepraia and Triatoma eratirusiformis. (A) cyt bgene sequences (514 bp) inferred with the model TrN +I, I: 0.66; (B) COI gene sequences (508 bp) model HKY+G, G: 0.068. Numbers above the branches are the support values derived from bootstrap resampling with 1000 pseudoreplicates and numbers under the branches are parsimony bootstrap values. Colors of nodes represent lineages. 21LA, 53Til in blue are the incongruent haplotypes of M. spinolai. Tera1, Tera2 in red are the incongruent haplotypes of T. eratyrusiformis. The bar represents the number of nucleotide substitutions per site.
Discussion
The extent of topological incongruence between two molecular markers depends both on evolutionary and methodological processes. For example, incongruence of mitochondrial and nuclear gene trees may be due to differential lineage sorting, introgression or heteroplasmy (Sota and Vogler, 2001; Magnacca and Brown, 2010; Barnabé and Brenière, 2012; Messenger et al., 2012). Different substitution rates might also lead to incongruence (Sasaki et al., 2005). However, due to the linked nature of mtDNA genes it is expected that different mitochondrial genes show similar topologies. Our phylogenetic analyses using two mtDNA genes (cyt b and COI) showed incongruent topologies that were evident for four haplotypes placed in different lineages. This unexpected result may be explained by: i) contamination of samples with foreign or neighboring DNA; ii) Taq polymerase may yield errors during the amplification process due to lack of fidelity; and iii) nucleotide intra-individual variation.
In this study, samples were extracted and amplified in different periods and were processed as groups of the same species, which rules out contamination errors. Similarly, it is highly unlikely that polymerase errors would be the primary source of the phylogenetic incongruence, as we used the same conditions for the Platinum® Taq DNA high fidelity polymerase in all our PCR reactions (Gyllensten, 1989; Frey and Frey, 2004). Hence, it is highly probable that the main cause of the incongruence is related to nucleotide intra-individual variation.
Nuclear integration of mitochondrial gene fragments has been reported in Triatominae (Dotson and Beard, 2001) and other arthropods (Zhang and Hewitt, 1996a; Parfait et al., 1998; Bensasson et al., 2001), but this process seems not to represent a major issue due to the larger number of mtDNA copies (Lightowlers et al., 1997; Scheffler, 2001). However, the presence of multiple nuclear copies of COI and other mitochondrial genes has been reported in aphids and other insects (Gellissen and Michaelis, 1987; Sunnucks and Hales, 1996; Bensasson et al., 2000), and therefore we do not rule out this phenomenon until further analyses are performed. These copies behave as pseudogenes that are not correctly expressed, because they accumulate nonsense mutations resulting in stop codons in the reading frame and a greater number of non-synonymous changes, thus tending to diverge from the homologous mitochondrial genes (Zhang and Hewitt, 1996a; Frey and Frey, 2004). We found no sequences with stop codons in the two genes, and found no non-synonymous mutations in the COI gene. Finally, the co-amplification of more than one mitochondrial haplotype (i.e. heteroplasmy; Solignac et al., 1986; Boyce et al., 1989), may explain our results. In the samples Tera1 Tera2 and 53Til we observed ambiguous bases after the nucleotide amplification (Figure 1); the sample 21LA did not show ambiguous bases.
The retention of ancestral polymorphisms through incomplete division of lineages may also explain the misplacement of the T. eratyrusiformis and M. spinolai haplotype. However, due to the linked nature of mitochondrial genes, the same results (incongruence) should have been observed in both topologies, contrary to our results. In addition, incomplete lineage sorting has a higher chance to occur in recently diverged groups and with high population sizes (Maddison and Knowles, 2006), which contrasts with the lineages in our study that are highly structured and show deep divergences (Campos et al., 2013a). Therefore, we suggest that heteroplasmy is the most parsimonious explanation for the incongruence observed in our study.
Heteroplasmy of mtDNA may result from two or more haplotypes within a single mitochondrion, or different mitochondria with different haplotypes, either within one cell or in different cells (Lightowlers et al., 1997; Frey and Frey, 2004). Processes that may explain the latter are paternal leakage, recombination, and segregating mutations (White et al., 2008). Paternal leakage or inheritance of mitochondrial DNA has been described in insects including Hemiptera (Kondo et al., 1990; Fontaine et al., 2007; Nunes et al., 2013; Wolff et al., 2013), and may have occurred between T. etratyrusiformis and Mepraia.This incongruence between mitochondrial genes due to heteroplasmy may lead to erroneous phylogenetic hypotheses resulting in wrong taxonomic classification and population genetic structure (Zhang and Hewitt, 1996b; White et al., 2008). The presumptive phenomenon of heteroplasmy suggests that mtDNA from T. eratyrusiformis can be found within the Mepriaia genome, which supports their close systematic relationship and is independent evidence of common ancestry. Due to the non-overlaping distribution of Mepraia and T. eratyrusiformis (separated by the Andes), the processes that may have caused the heteroplasmy seen within the spinolai complex probably occurred a long time ago, before or at early periods during the uplifting of the Andes Ranges, and reveal that heteroplasmy may persist in populations even though new lineages are subsequently formed.
In conclusion, after performing phylogenetic reconstructions with two mitochondrial genes, we found incongruent topologies for some haplotypes of M. spinolai and T. eratyrusiformis. We suggest as explanation of our results that there is intra-individual variation likely due to heteroplasmy. The mitochondrial incongruence within the spinolaicomplex requires further investigation to determine more accurately the extent and probable causes of this pattern.
Acknowledgments
We thank Carezza Botto-Mahan and S.S. Catalá for providing valuable kissing-bug samples. This work was supported by funds from CONICYT program FONDECYT 3150289, the postdoctoral project 2014 VRIEA PUCV (R. Campos) and FONDECYT 1085154 (A. Solari).
Footnotes
Associate Editor: Fabrício Rodrigues dos Santos
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Autom Contr. 1974;19:716–723. [Google Scholar]
- Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC. Intraspecific phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annu Rev Ecol Syst. 1987;18:489–522. [Google Scholar]
- Barnabé C, Brenière SF. Scarce events of mitochondrial introgression in Trypanosoma cruzi: New case with a Bolivian strain. Infect Genet Evol. 2012;12:1879–1883. doi: 10.1016/j.meegid.2012.08.018. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang DX, Hewitt GM. Frequent assimilation of mitochondrial DNA by grasshopper nuclear genomes. Mol Biol Evol. 2000;17:406–415. doi: 10.1093/oxfordjournals.molbev.a026320. [DOI] [PubMed] [Google Scholar]
- Bensasson D, Zhang DX, Hartl DL, Hewitt GM. Mitochondrial pseudogenes: Evolution’s misplaced witnesses. Trends Ecol Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- Botto-Mahan C, Sepúlveda M, Vidal M, Acuña-Retamar M, Ortiz S, Solari A. Trypanosoma cruzi infection in the sylvatic kissing bug Mepraia gajardoi from the Chilean Southern Pacific Ocean coast. Acta Trop. 2008;105:166–169. doi: 10.1016/j.actatropica.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Boyce TM, Zwick ME, Aquadro CF. Mitochondrial DNA in the bark weevils: Size, structure and heteroplasmy. Genetics. 1989;123:825–836. doi: 10.1093/genetics/123.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleros L, Panzera F, Bargues MD, Monteiro FA, Klisiowicz DR, Zuriaga MA, Mas-Coma S, Pérez R. Systematics of Mepraia (Hemiptera-Reduviidae): Cytogenetic and molecular variation. Infect Genet Evol. 2010;10:221–228. doi: 10.1016/j.meegid.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Campos R, Botto-Mahan C, Coronado X, Jaramillo N, Panzera F, Solari A. Wing shape differentiation of Mepraia species (Hemiptera, Reduviidae) Infect Genet Evol. 2011;11:329–333. doi: 10.1016/j.meegid.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Campos R, Torres-Pérez F, Botto-Mahan C, Coronado X, Solari A. High phylogeographic structure in sylvatic vectors of Chagas disease of the genus Mepraia (Hemiptera, Reduviidae) Infect Genet Evol. 2013a;19:280–286. doi: 10.1016/j.meegid.2013.04.036. [DOI] [PubMed] [Google Scholar]
- Campos R, Botto-Mahan C, Coronado X, Catala SS, Solari A. Phylogenetic relationships of the spinolai complex and other Triatomini based on mitochondrial DNA sequences (Hemiptera, Reduviidae) Vector Borne Zoonotic Dis. 2013b;13:73–76. doi: 10.1089/vbz.2011.0954. [DOI] [PubMed] [Google Scholar]
- Cattan PE, Pinochet A, Botto-Mahan C, Acuña MI, Canals M. Abundance of Mepraia spinolai in a periurban zone of Chile. Mem Inst Oswaldo Cruz. 2002;97:285–287. doi: 10.1590/s0074-02762002000300001. [DOI] [PubMed] [Google Scholar]
- De Paula AS, Diotaiuti L, Schofield CJ. Testing the sister-group relationship of the Rhodniini and Triatomini (Insecta, Hemiptera, Reduviidae, Triatominae) Mol Phylogenet Evol. 2005;35:712–718. doi: 10.1016/j.ympev.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Dos K, Paduan S, Eduardo P, Ribolla M. Mitochondrial DNA polymorphism and heteroplasmy in populations of Aedes aegypti in Brazil. J Med Entomol. 2008;45:59–67. doi: 10.1603/0022-2585(2008)45[59:mdpahi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Dotson EM, Beard CB. Sequence and organization of the mitochondrial genome of the Chagas disease vector, Triatoma dimidiata . Insect Mol Biol. 2001;10:205–215. doi: 10.1046/j.1365-2583.2001.00258.x. [DOI] [PubMed] [Google Scholar]
- Dujardin JP, Panzera P, Schofield CJ. Triatominae as a model of morphological plasticity under ecological pressure. Mem Inst Oswaldo Cruz. 1999;94:223–228. doi: 10.1590/s0074-02761999000700036. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Fontaine KM, Cooley JR, Simon C. Evidence for paternal leakage in hybrid periodical cicadas (Hemiptera, Magicicada spp.) PLoS One. 2007;2:e892. doi: 10.1371/journal.pone.0000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey JE, Frey B. Origin of intra-individual variation in PCR-amplified mitochondrial cytochrome oxidade I of Thrips tabaci(Thysanoptera, Thripidae): Mitochondrial heteroplasmy or nuclear integration? Hereditas. 2004;140:92–98. doi: 10.1111/j.1601-5223.2004.01748.x. [DOI] [PubMed] [Google Scholar]
- Frías D. A new species and karyotype variation in the bordering distribution of Mepraia spinolai (Porter) and Mepraia gajardoi Frías et al. (Hemiptera, Reduviidae, Triatominae) in Chile and its parapatric model of speciation. Neotrop Entomol. 2010;39:572–583. doi: 10.1590/s1519-566x2010000400017. [DOI] [PubMed] [Google Scholar]
- Frías D, Henry A, Gonzalez C. Mepraia gajardoi: A new species of tritominae (Hemiptera, Reduviidae) from Chile and its comparison with Mepraia spinolai . Rev Chil Hist Nat. 1998;71:177–188. [Google Scholar]
- Gellissen G, Michaelis G. Gene transfer: Mitochondria to nucleus. Ann NY Acad Sci. 1987;503:391–401. doi: 10.1111/j.1749-6632.1987.tb40625.x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. Direct sequencing of in vitro amplified DNA. In: Ehrlich HA, editor. PCR Technology Principles and Applications for DNA Amplification. Stockton P; London: 1989. pp. 45–60. [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- Hillis DM, Bull JJ, Url S. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol. 1993;42:182–192. [Google Scholar]
- Hypa V, Tietz DF, Zrzavy J, Rego ROM, Galvao C, Jurberg J. Phylogeny and biogeography of Triatominae (Hemiptera, Reduviidae): Molecular evidence of a New World origin of the Asiatic clade. Mol Phylogenet Evol. 2002;23:447–457. doi: 10.1016/s1055-7903(02)00023-4. [DOI] [PubMed] [Google Scholar]
- Justi SA, Russo CAM, Mallet JRDS, Obara MT, Galvão C. Molecular phylogeny of Triatomini (Hemiptera, Reduviidae, Triatominae) Parasit Vectors. 2014;7:149. doi: 10.1186/1756-3305-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo R, Satta Y, Matsuura ET, Ishiwa H, Takahata N, Chigusa SI. Incomplete maternal transmission of mitochondrial DNA in Drosophila . Genetics. 1990;126:657–663. doi: 10.1093/genetics/126.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lent H, Wygodzinsky P. Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas disease. Bull Am Mus Nat Hist. 1979;163:123–520. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lightowlers RN, Chinnery PF, Turnbull DM, Howell N, Turnbuu DM. Mammalian mitochondrial genetics: Heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–455. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Knowles LL. Inferring phylogeny despite incomplete lineage sorting. Syst Biol. 2006;55:21–30. doi: 10.1080/10635150500354928. [DOI] [PubMed] [Google Scholar]
- Magnacca KN, Brown MJ. Mitochondrial heteroplasmy and DNA barcoding in Hawaiian Hylaeus (Nesoprosopis) bees (Hymenoptera, Colletidae) BMC Evol Biol. 2010;10:e174. doi: 10.1186/1471-2148-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza S, Gajardo R, Jörg M. Mepraia novum genus de Triatominae. Mepraia spinolai (Porter) 1933, redescripción del macho y descripción de la hembra. MEPRA Publicación. 1940;44:3–30. [Google Scholar]
- Messenger LA, Llewellyn MS, Bhattacharyya T, Franzén O, Lewis MD, Ramírez JD, Carrasco HJ, Andersson B, Miles MA. Multiple mitochondrial introgression events and heteroplasmy in Trypanosoma cruzi revealed by maxicircle MLST and next generation sequencing. PLoS Negl Trop Dis. 2012;6:e1584. doi: 10.1371/journal.pntd.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro FA, Barrett TV, Fitzpatrick S, Cordon-Rosales C, Feliciangeli D, Beard CB. Molecular phylogeography of the Amazonian Chagas disease vectors Rhodnius prolixus and R. robustus . Mol Ecol. 2003;12:997–1006. doi: 10.1046/j.1365-294x.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- Moreno ML, Gorla D, Catalá S. Association between antennal phenotype, wing polymorphism and sex in the genus Mepraia (Reduviidae, Triatominae) Infect Genet Evol. 2006;6:228–234. doi: 10.1016/j.meegid.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Nunes MDS, Dolezal M, Schlötterer C. Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster . Mol Ecol. 2013;22:2106–17. doi: 10.1111/mec.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfait B, Rustin P, Munnich A, Rötig A. Co-amplification of nuclear pseudogenes and assessment of heteroplasmy of mitochondrial DNA mutations. Biochem Biophys Res Commun. 1998;247:57–59. doi: 10.1006/bbrc.1998.8666. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rozas M, Botto-Mahan C, Coronado X, Ortiz S, Cattan PE, Solari A. Coexistence of Trypanosoma cruzi genotypes in wild and periodomestic mammals in Chile. Am J Trop Med Hyg. 2007;77:647–653. [PubMed] [Google Scholar]
- Sasaki T, Nikaido M, Hamilton H, Goto M, Kato H, Kanda N, Pastene L, Cao Y, Fordyce R, Hasegawa M, Okada N. Mitochondrial phylogenetics and evolution of mysticete whales. Syst Biol. 2005;54:77–90. doi: 10.1080/10635150590905939. [DOI] [PubMed] [Google Scholar]
- Scheffler IE. A century of mitochondrial research: Achievements and perspectives. Mitochondrion. 2001;1:3–31. doi: 10.1016/s1567-7249(00)00002-7. [DOI] [PubMed] [Google Scholar]
- Solignac M, Monnerot M, Mounolou JC. Mitochondrial DNA evolution in the melanogaster species subgroup of Drosophila . J Mol Evol. 1986;23:31–40. doi: 10.1007/BF02100996. [DOI] [PubMed] [Google Scholar]
- Sota T, Vogler AP. Incongruence of mitochondrial and nuclear gene trees in the Carabid beetles Ohomopterus . Syst Biol. 2001;50:39–59. [PubMed] [Google Scholar]
- Sunnucks P, Hales DF. Numerous transposed sequences of mitochondrial cytochrome oxidase I–II in aphids of the genus Sitobion (Hemiptera, Aphididae) Mol Biol Evol. 1996;13:510–524. doi: 10.1093/oxfordjournals.molbev.a025612. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony * (and other methods) version 40b10. Sinauer Associates; Sunderland: 2002. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo A, Vergara F, Campos R, Botto-Mahan C, Ortiz S, Coronado X, Solari A. Trypanosoma cruzi genotypes in Mepraia gajardoi from wild ecotopes in northern Chile. Am J Trop Med Hyg. 2013;88:285–288. doi: 10.4269/ajtmh.2012.12-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DJ, Wolff JN, Pierson M, Gemmell NJ. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol. 2008;17:4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x. [DOI] [PubMed] [Google Scholar]
- Wilcox TP, Zwickl DJ, Heath TA, Hillis DM. Phylogenetic relationships of the dwarf boas and a comparison of Bayesian and bootstrap measures of phylogenetic support. Mol Phylogenet Evol. 2002;25:361–371. doi: 10.1016/s1055-7903(02)00244-0. [DOI] [PubMed] [Google Scholar]
- Wolff JN, Nafisinia M, Sutovsky P, Ballard JWO. Paternal transmission of mitochondrial DNA as an integral part of mitochondrial inheritance in metapopulations of Drosophila simulans . Heredity. 2013;110:57–62. doi: 10.1038/hdy.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DX, Hewitt GM. Nuclear integrations: Challenges for mitochondrial DNA markers. Trends Ecol Evol. 1996a;11:247–251. doi: 10.1016/0169-5347(96)10031-8. [DOI] [PubMed] [Google Scholar]
- Zhang DX, Hewitt GM. Highly conserved nuclear copies of the mitochondrial control region in the desert locust Schistocerca gregaria: Some implications for population studies. Mol Ecol. 1996b;5:295–300. doi: 10.1111/j.1365-294x.1996.tb00317.x. [DOI] [PubMed] [Google Scholar]
- Zhang DX, Szymura JM, Hewitt GM. Evolution and structural conservation of the control region of insect mitochondrial DNA. J Mol Evol. 1995;40:382–391. doi: 10.1007/BF00164024. [DOI] [PubMed] [Google Scholar]
Internet Resources
- FigTree v1.1.2 program. [Mar 20;2013 ]. http://tree.bio.ed.ac.uk/software/figtree/