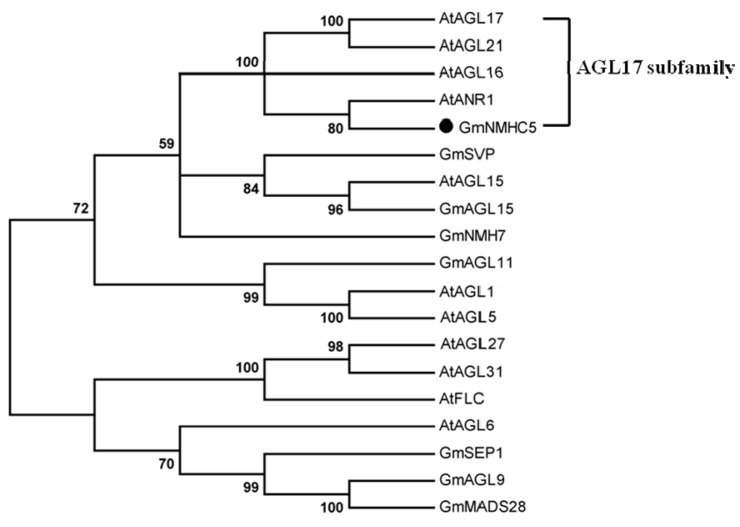
Figure 2.
Phylogenetic tree based on protein sequences between GmNMHC5 and some other function-known MIKC-type MADS-box transcription factors of Glycine max and Arabidopsis thaliana (Gm, G. max; At, Arabidopsis thaliana). Accession numbers are as follows: AtAGL17 (NP_179848.1), AtAGL21 (NP_195507.1), AtAGL16 (NP_191282.2), AtANR1 (CAB09793.1), GmNMHC5 (NP_001241489.1), GmSVP (ACJ61500.1), AtAGL15 (NP_196883.1), GmAGL15 (NP_001237033.1), GmNMH7 (NP_001236857.1), GmAGL11 (NP_001236130.1), GmAGL1 (NP_191437.1), GmAGL5 (NP_565986.1), GmAGL27 (NP_177833.3), AtAGL31 (NP 001119498.1), AtFLC (NP_196576.1), AtAGL6 (NP_182089.1), GmSEP1 (NP_001238296.1), GmAGL9 (ACA24481.1), GmMADS28 (NP 001236390.1). The Phylogenetic tree was constructed using the Maximum Likelihood method of phylogenetic tree construction, with 200 bootstrap replicates, using MEGA v.5.05. The number for each node is the bootstrap percentages, and nodes with less than 70% bootstrap values were collapsed.