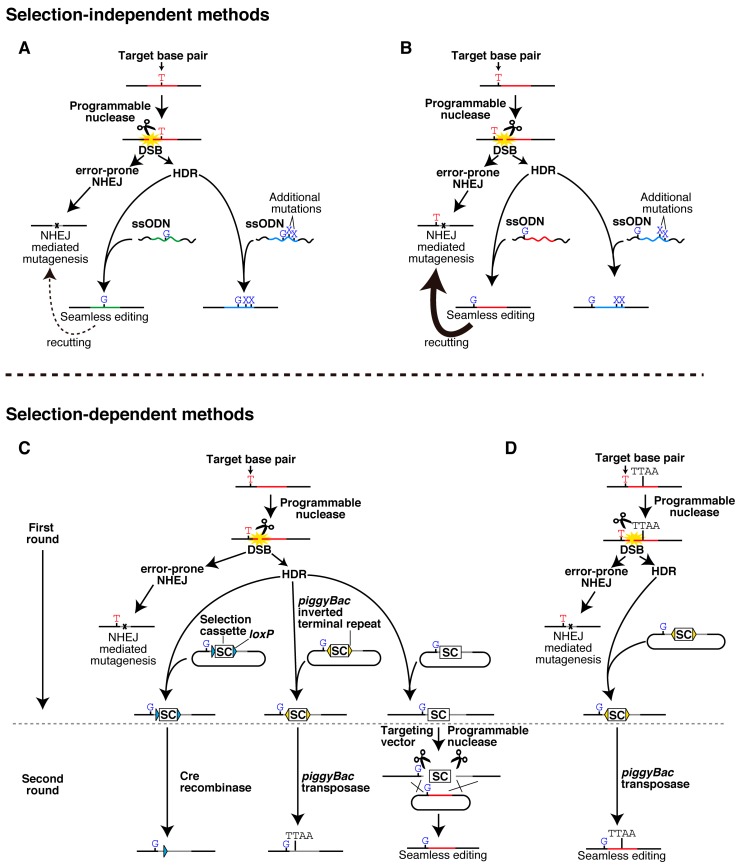
Figure 1.
Single-base pair genome editing and related techniques. Schematic representation of methods to engineer a single or a small number of nucleotide substitutions into the genome. (A,B) Selection-independent editing of a target base pair within (A) and out (B) of a sequence recognized by a programmable nuclease; (C,D) Selection-dependent editing using different methods to excise the selection marker. piggyBac-excision is illustrated without (C) or with (D) a TTAA sequence naturally present near or at the target site. Blue, gray, red, and green lines represent programmable nuclease target sites with deletions, insertions, single, and multiple nucleotide substitutions, respectively. Target sites with single nucleotide substitutions may be recut by the programmable nuclease depending on the properties of the enzyme and the location of the substitution. Scissors and yellow star shapes represent programmable endonucleases and DNA double-strand breaks (DSBs), respectively.