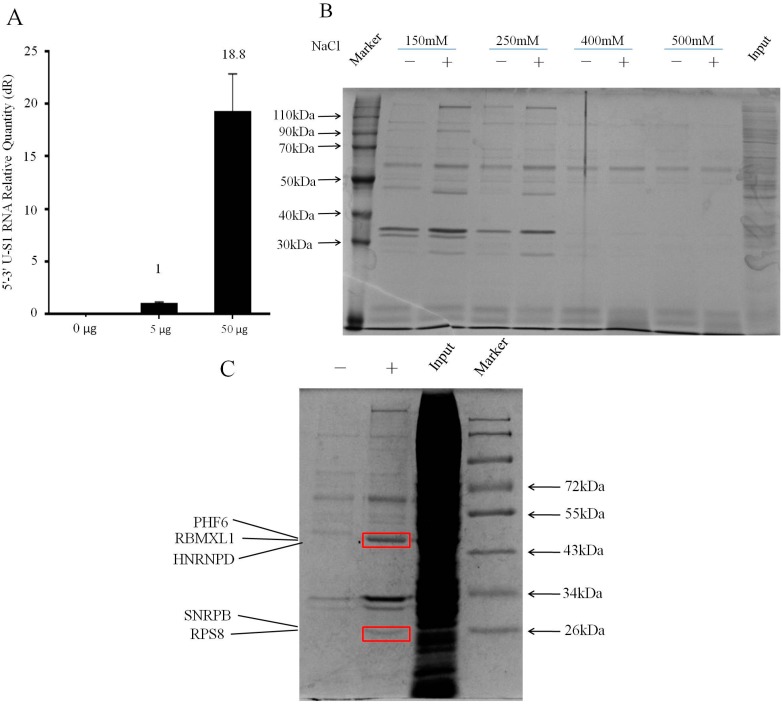
Figure 2.
Purification of endogenously-assembled 5′–3′ U-S1 RNPs. (A) The 5′–3′ U-S1RNAs eluted from streptavidin beads were detected by RT-qPCR with different amount of plasmid shown. In order to normalize the RT-qPCR, an equal amount of β-actin RNA was added to both groups prior to isolating RNA from beads; (B) In the RNA pull down assay, the wash buffer containing different NaCl concentrations was applied to explore the perfect wash conditions for affinity purification. Proteins bound to washed streptavidin beads from 5′–3′ U-S1 RNA and mock purifications were resolved by SDS-PAGE and visualized by Coomassie blue staining. Lanes marked with a minus are untransfected controls; (C) The two distinguished gel bands of the 5′–3′ U-S1 RNA purification compared to mock purification were subjected to mass spectrometry, and five proteins were identified.