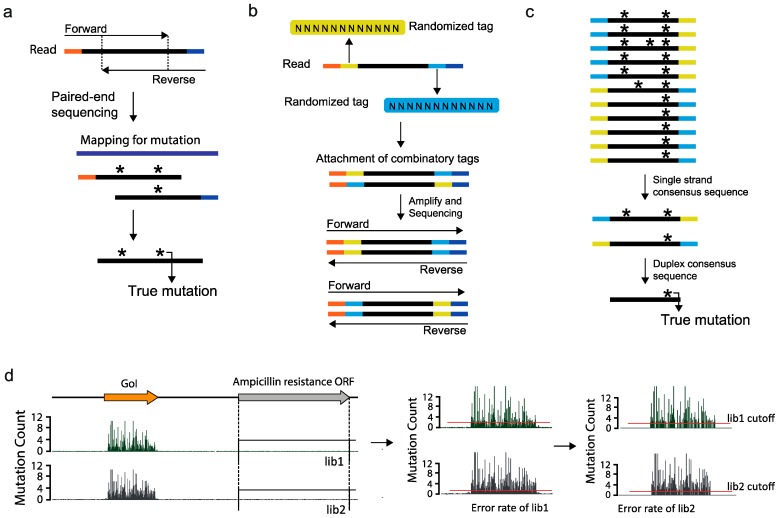
Figure 3.
Methods to rectify sequencing errors: (a) a scheme of how paired end reads with short sequencing reads allow the detection of sequencing errors; (b) schematic showing the concept of the duplex sequencing method; (c) how the consensus sequence is used to remove sequencing errors, adapted by permission from the Macmillan Publishers Ltd: Nature Protocols [61], copyright 2014. The black bar indicate the target inserts reads for sequencing and the orange and dark blue colored bars at end of the insert reads indicate sequencing adaptors; the yellow and light blue bars indicate the randomized duplex tags; and (d) Hypothetical mapping of the mutation frequency for variant library sequencing. The red line indicates the sequencing error rate of the ampicillin gene used as the cutoff. The asterisks indicate mutations and the orange and blue bars at the ends of the reads indicate the sequencing adaptors.