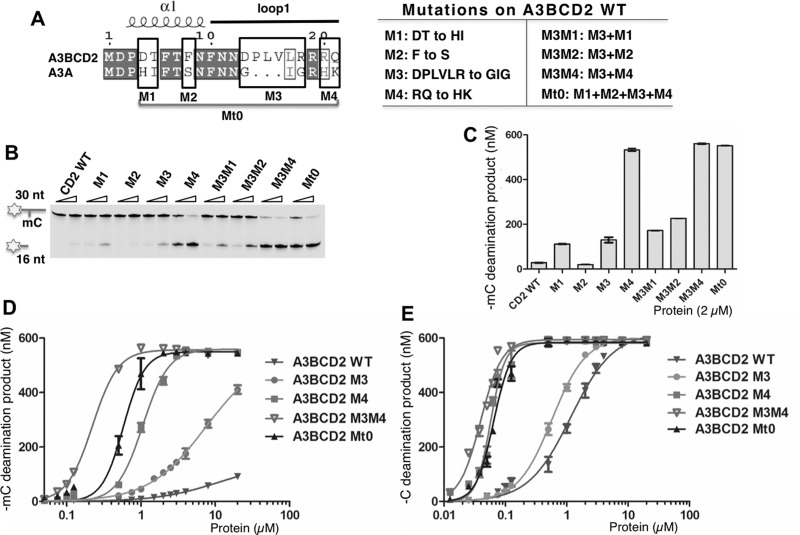
Figure 5. Dissecting loop-1 residues important for increasing mC deamination.
Error bars represent S.D. from the mean for three independent experiments. (A) Design of the mutants on A3BCD2 WT. Sequence alignment of A3BCD2 and A3A showed four groups (M1–M4) around loop-1 that are not conserved (left). A3BCD2 was mutated to contain the corresponding residues of A3A individually to generated mutant M1–M4. A3BCD2 M3 was combined with others to generate three additional combined mutants. (B) Gel image showing the different mC deamination activities by the mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (C) Quantification of the activity on mC. The activity of the mutants at the concentration of 2 μM was quantified. (D) Dose-response of mC deamination activity by A3BCD2 WT, M3, M4, M3M4 and Mt0 constructs. (E) Dose-response of C deamination activity by A3BCD2 WT, M3, M4, M3M4 and Mt0 constructs.