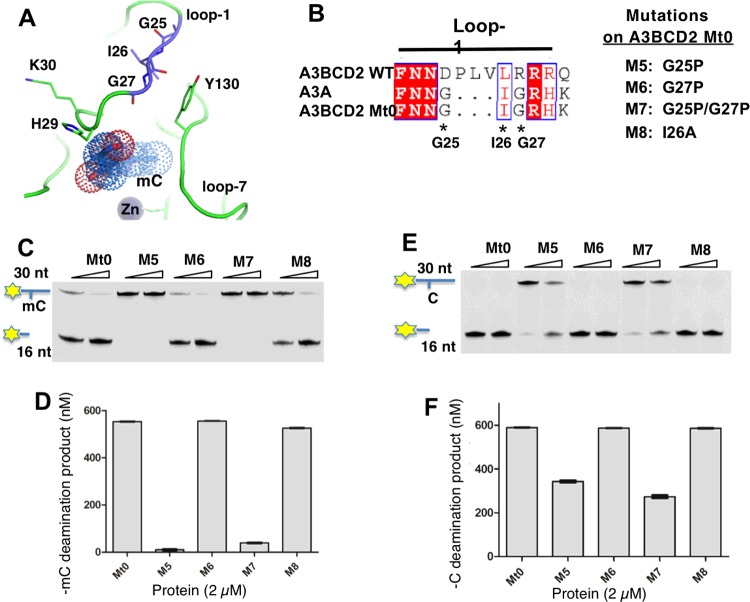
Figure 6. The flexibility of loop-1 conformation in A3BCD2 Mt0 affects mC deamination activity.
Error bars represent S.D. from the mean for three independent experiments. (A) Structure of A3A loop-1 and Tyr130 on loop-7 around the Zn active site. Residues–G25I26G27–and–H29K30–on loop-1 are drawn as sticks. The mC modelled into the active site is shown as dots. In the NMR structure of A3A (PDB: 2M65), Tyr130 is close to–GIG–. (B) Design of four mutants on loop-1 of A3BCD2 Mt0. (C) Gel image showing the mC deamination activity of the mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (D) Quantification of the mC deamination activity at a protein concentration of 2 μM. (E) Gel image showing the C deamination activity of the mutants. Each protein (0.5 μM and 2 μM) was incubated with 600 nM 30 nt ssDNA substrates at 37°C for 2 h. (F) Quantification of the C deamination activity at a protein concentration of 2 μM.