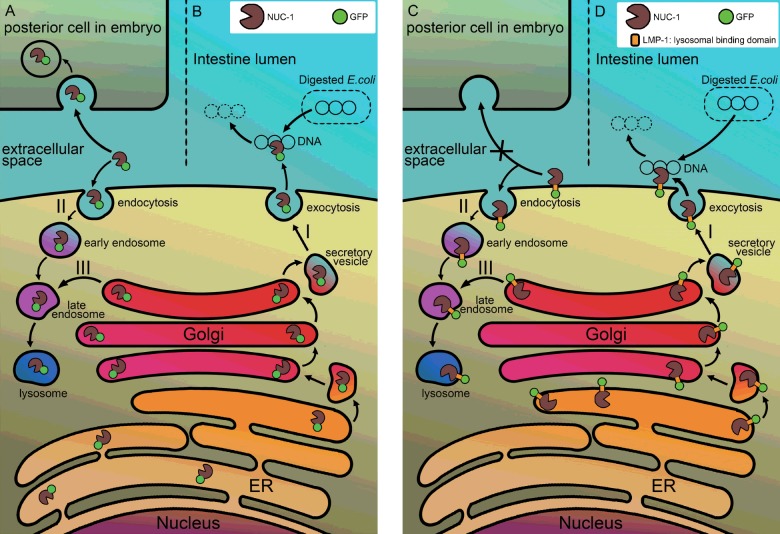
Figure 8. A hypothetical model illustrates the maturation pathway and action mode of two NUC-1 fusion proteins.
The N-terminal leader sequence present in the NUC-1 of fusion proteins leads the polypeptide to ER for completion of biosynthesis. The NUC-1::GFP protein is then released into the cisternae of ER as a free form of protein in smIs172 animals (left panel). The additional LMP-1 sequence present in the NUC-1::LMP-1153–237::GFP protein renders the fusion protein appearing as a transmembrane protein, in which the GFP is located outside of ER and the DNase II domain is facing to ER lumen (right panel). Most of fusion proteins are resided in ER, upon signalling of bacterial DNA or apoptosis, both move to Golgi apparatus for further glycosylation and being secreted through vesicles transportation to the plasma membrane. After the vesicles are fused with plasma membrane, the NUC-1::GFP fusion protein is released into the lumen of intestine for degradation of bacterial DNA (B) or into the extracellular space and then being retaken up by neighbouring cells (A) for degradation of apoptotic DNA (pathway I). The secreted fusion protein can also go through endocytosis mechanism back to lysosomes of cells (pathway II). Whereas the NUC-1::LMP-1153–237::GFP protein remains on the plasma membrane of intestine cells or embryonic blastomeres (D) (pathway I), it is able to digest bacterial DNA but unable to be retaken up by posterior cells for degradation of apoptotic DNA non-autonomously (C). However, it can be recycled through endocytosis pathway to target to lysosomes (pathway II). Both fusion proteins have a regular pathway targeting to lysosomes (pathway III). Symbols of fusion proteins are indicated by green circle as GFP and a pacman as DNase II.