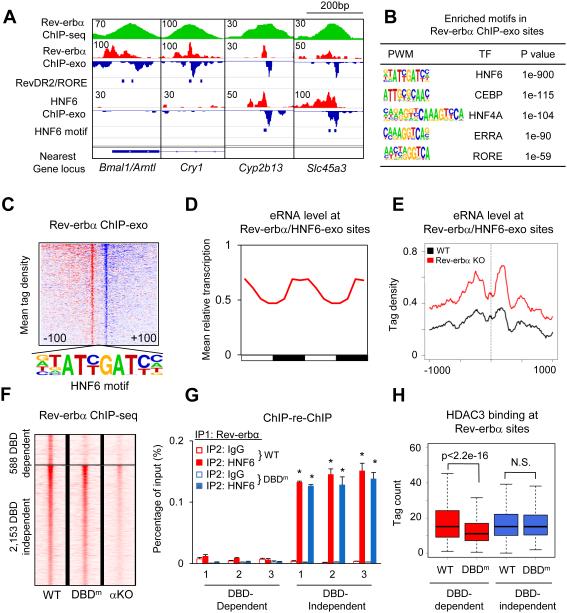
Figure 2. Rev-erbα binds to the genome using both DBD-dependent and DBD-independent mechanism.
(A) Genome browser view of Rev-erbα and HNF6 ChIP-exo signals under Rev-erbα ChIP-seq peaks near clock and metabolic genes. Blue bars indicate locations of RevDR2/RORE and HNF6 motif. (B) Highly enriched known motifs found in Rev-erbα ChIP-exo peak pairs 22-26bp apart. (C) Heat map showing 5’-end tag densities of Rev-erbα ChIP-exo centered at HNF6 motifs within 1,108 peak pairs. Red and blue indicate tag density on the plus and minus strand, respectively. (D) Mean relative eRNA transcription (22) at Rev-erbα/HNF6-exo sites throughout 24-hour light dark cycles. Data were double plotted for clearer visualization. (E) eRNA tag density (22) centered at Rev-erbα/HNF6-exo sites near Rev-erbα target genes, in Rev-erbα KO and wild type liver. (F) Heat map showing Rev-erbα ChIP-seq tag densities (at ZT10) in wild type, DBD mutant (DBDm), and Rev-erbα KO (αKO) mice, at DBD-dependent and -independent sites identified among 5,792 high confidence Rev-erbα peaks (peak height >1rpm, WT/Rev-erbα KO>3). (G) Sequential ChIP of Rev-erbα followed by either HNF6 or IgG ChIP in wild type and DBDm mouse liver at ZT10. Data are expressed as mean± SEM (* Student’s t-test, p<0.05, n=3 or 4 per group). (H) Binding of HDAC3 at DBD-dependent and -independent sites in WT and DBDm liver (N.S. Student’s t-test, p>0.05).