Table 1. Optimization of the Reaction Parametersa.
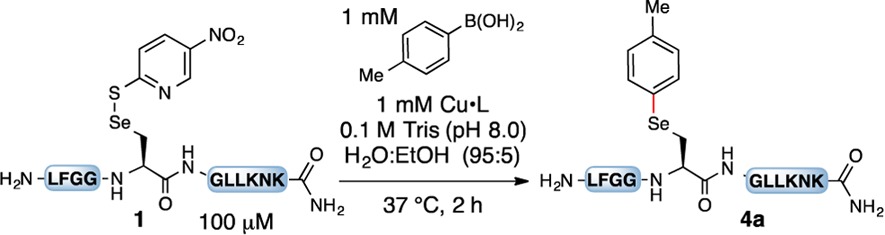
| entry | Cu | ligand | % yield (% conversion)b |
|---|---|---|---|
| 1 | CuSO4 | L1 | 79 (99) |
| 2 | CuSO4 | L2 | 81 (99) |
| 3 | CuSO4 | L3 | 66 (99) |
| 4 | CuSO4 | L4 | 17 (91) |
| 5 | CuSO4 | – | 5 (30) |
| 6 | CuCl2·2H2O | L2 | 78 (99) |
| 7 | CuBr2 | L2 | 79 (99) |
| 8 | Cu(OAc)2 | L2 | 78 (99) |
| 9 | – | L2 | 0 (22)c |
| 10d | CuSO4 | L2 | 44 (72) |
| 11e | CuSO4 | L2 | 7 (30) |
| 12f | CuSO4 | L2 | NR |
| 13g | CuSO4 | L2 | 5 (99) |
| 14h | CuSO4 | L2 | 4 (99) |
| 15i | CuSO4 | L2 | NR |
See the Supporting Information for details. Amino acids are shown in a one-letter code. NR = no reaction.
Yields were determined by integration of the total ion currents (TICs) from LC–MS analyses of the unpurified reaction mixtures.
Elimination and diselenide were the only observable products.
0.5 mM CuSO4, 0.5 mM L2, and 0.5 mM boronic acid were used.
0.25 mM CuSO4, 0.25 mM L2, and 0.25 mM boronic acid were used.
The Sec-TNP residue replaced with Ser.
The Sec-TNP residue was replaced with Cys.
The Sec-TNP residue was replaced with Cys-TNP.
The Sec-TNP residue was replaced with Met.
