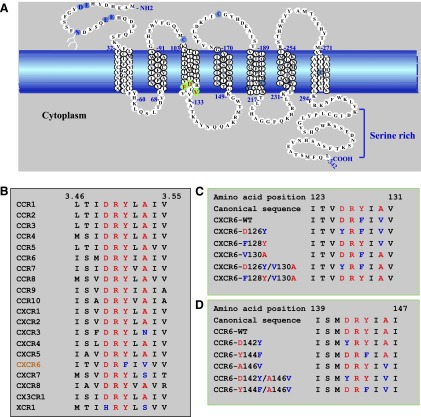
Fig. 1.
The sequence of CXCR6 has atypical features. (A) Schematic representation of the wild-type CXCR6 receptor. The cell membrane is represented by the blue band. Amino acids in transmembrane α helices are shown in stacked triplets. CXCR6 contains residues, shown in light blue, that are conserved in GPCRs, such as N-linked glycosylation site(s) in the amino-terminal domain (N16 with branched structure), cysteines in extracellular loops one and two (C102 and C180), as well as sequences that are characteristic of chemokine receptors, such as paired acidic residues in the amino-terminal domain (E8 and D9 and E21 and E22), a paired cysteine and tyrosine (C210 and Y211), and a cysteine in helix seven (C282). Residues that we changed by site-directed mutagenesis are indicated in green with red letters. (B) Amino acid sequence alignment of H3C in CXCR6 and other chemokine receptors. Residue numbers 3.46 and 3.55 are according to the convention of Ballesteros and Weinstein (1995) (see text). (C) Sequences of CXCR6 wild type and mutants that we produced and studied. Numbers designate CXCR6 I123 and V131. Canonical residues are red, and noncanonical residues are blue. (D) Sequences of CCR6 wild type and mutants that we produced and studied. Numbers designate CCR6 I139 and I147. Canonical residues are red, and noncanonical residues are blue.