Fig. 4.
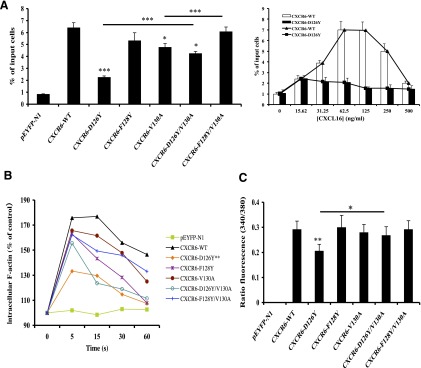
CXCR6-D126Y has impaired activity when expressed in Jurkat E6-1 cells. (A) Cells were transfected with variable amounts of DNAs encoding wild-type and mutant CXCR6-YFP receptors to produce equal surface expression. In each experiment, the means were obtained for the percentage of input cells migrating to duplicate wells of a Transwell plate, as described in Materials and Methods. The left panel shows mean ± S.E.M. of data from three experiments. The asterisks indicate a significant difference from Jurkat E6-1 cells expressing the wild-type receptor or between the cells connected by the horizontal bars. The right panel shows mean ± S.E.M. of data from three experiments using cells transfected with wild-type CXCR6 DNA or CXCR6-D126Y DNA and lower wells containing various concentrations of CXCL16. (B) Cells transfected with equal amounts of DNA encoding wild-type and mutant CXCR6-YFP proteins were fixed and stained with Alexa Fluor 488 phalloidin-paraformaldehyde at the indicated times after stimulation with 2.5 μg/ml CXCL16, as described in Materials and Methods. All results are plotted relative to the mean fluorescence intensity of the cells without the addition of CXCL16, which was set at 100% and plotted at 0 seconds. The mean responses at each time point were calculated from the three experiments, and the asterisks indicate significant differences between the curves for D126Y versus wild-type receptors. (C) Cells transfected as in (A) were loaded with Fura-2/AM and assayed for intracellular calcium mobilization on a spectrofluorometer in response to 1 μg/ml CXCL16, as described in Materials and Methods. Mean peak responses ± S.E.M. were calculated from five experiments, and the asterisks indicate significant differences between D126Y versus wild-type receptors and, as indicated by the bar, mutants D126Y versus D126Y/V130A. *P < 0.05; **P < 0.01; ***P < 0.001.
