Figure 2.
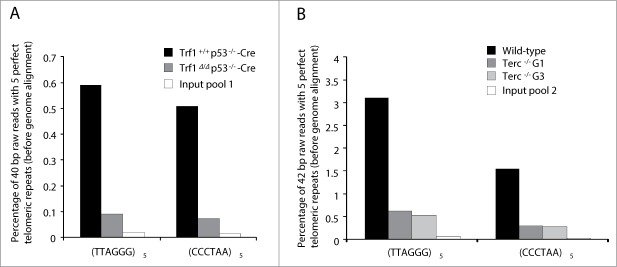
Enrichment of telomere sequences bound to TRF1 in ChIP-seq experiments. The percentage of 40 or 42-bp raw reads before alignment with mouse genome containing perfect (TTAGGG)5 or (CCCTAA)5 repeats is shown for the different samples. In the first experiment, input pool 1 is a combination of equal amounts of input DNA (total DNA before immunoprecipitation) from Trf1+/+ p53−/−-Cre and Trf1Δ/Δ p53−/−-Cre samples (A). In the second experiment, input pool 2 is a combination of input DNA from wild-type, Terc−/− G1 and Terc−/− G3 samples (B).
