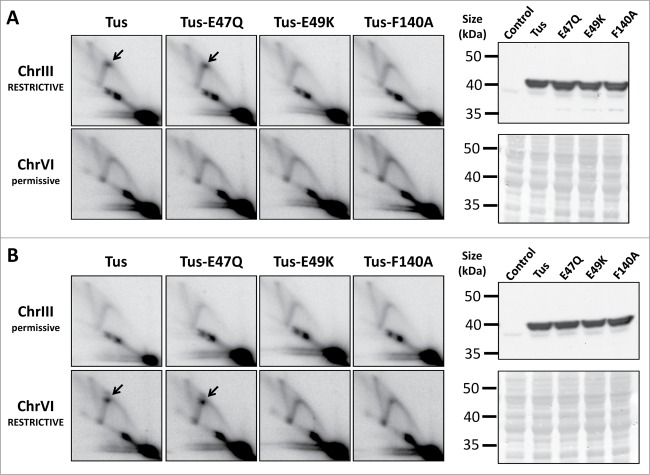
Figure 1.
Mutational analysis of the Tus-Ter complex when reconstituted in S. cerevisiae. Yeast strains engineered with either restrictive (blocking) or permissive (non-blocking) 3xTerB modules adjacent to ARS305 (on ChrIII) and ARS607 (on ChrVI) were transformed with a low-copy GAL1-regulated plasmid containing HA-tagged wild-type Tus, or HA-Tus with E47Q, E49K, or F140A substitutions. The 3xTerB modules examined here were arranged in either (A) the ChrIII RESTRICTIVE / ChrVI permissive orientation, or (B) the reciprocal ChrIII permissive / ChrVI RESTRICTIVE configuration. At both of these genomic loci, replication forks emanating from either ARS305 or ARS607 replicate these 3xTerB modules from left to right. Cultures were synchronized in G1 with α-factor pheromone, and expression of Tus was induced for 2.5 hours during the cell synchronization step. Following a 35 min release from G1-arrest, cells were harvested for 2D gel analysis (left panels), or Western blotting for HA-Tus (right panels). The 2D gel images show DNA replication intermediates detectable at (a BamHI-HindIII fragment of) ChrIII and (a HindIII-HindIII fragment of) ChrVI restriction fragments. Paused RFs at Tus-Ter are indicated by the black arrow. Western blotting confirmed equivalent levels of wild type and mutated HA-Tus proteins at this time point. A control (uninduced HA-Tus) sample was also included as a negative control for the Western blot analysis. Membranes were stained with Ponceau S to confirm equivalent protein loading (lower right panels).