The Hippo pathway is an emerging tumor suppressor pathway that was initially discovered by elegant genetic studies performed in Drosophila. It is now known that this pathway restrains organ size by stimulating programmed cell death and limiting excessive cell growth and division, all properties critical for tissue homeostasis and tumor suppression.
In mammalian cells, the core of the Hippo pathway is composed of 2 serine/threonine kinases, MST1/2 and LATS1/2. Once activated, MST phosphorylates the scaffold proteins SAV1 and MOB1, as well as LATS. This is postulated to lead to activation of LATS and in turn phosphorylation of YAP (Yes-associated protein). YAP is the key downstream effector in the Hippo pathway, serving as a transcriptional co-activator that regulates tissue growth by controlling the expression of many proliferation and anti-apoptosis—related genes. Phosphorylation of YAP by LATS leads to its retention in the cytoplasm by 14-3-3 and therefore inhibition of its transactivation activity. Although a number of regulators of YAP have been identified through many biochemical and genetic studies, the upstream pathways and extracellular signals that control the mammalian Hippo pathway remain unclear.
We and Dr. Guan's lab have recently reported energy homeostasis as a critical regulating event controlling activation of the Hippo pathway.1,2 YAP activity is robustly suppressed under energy stress conditions, such as glucose starvation or treatment with 2-deoxy-D-glucose (2-DG). Both the LATS kinase and AMP-activated protein kinase (AMPK) are activated under energy stress, phosphorylating YAP and contributing to its inactivation. LATS kinase phosphorylates YAP at Ser127, which promotes YAP nucleus-to-cytoplasm translocation. AMPK phosphorylates YAP at several sites, among which Ser94 and Ser61 sites are the most important ones. The phosphorylation of Ser94 disrupted the association between YAP and TEAD, which suppressed their transcriptional activity.1,2 The phosphorylation of Ser61 dramatically reduced the AMPK-mediated YAP migration shift in phospho-tag gel and inhibited YAP activity through an unknown mechanism.1 These studies demonstrate energy stress to be an upstream signal that controls the Hippo pathway and reveal AMPK as a kinase that directly phosphorylates and regulates YAP (Fig. 1).
Figure 1.
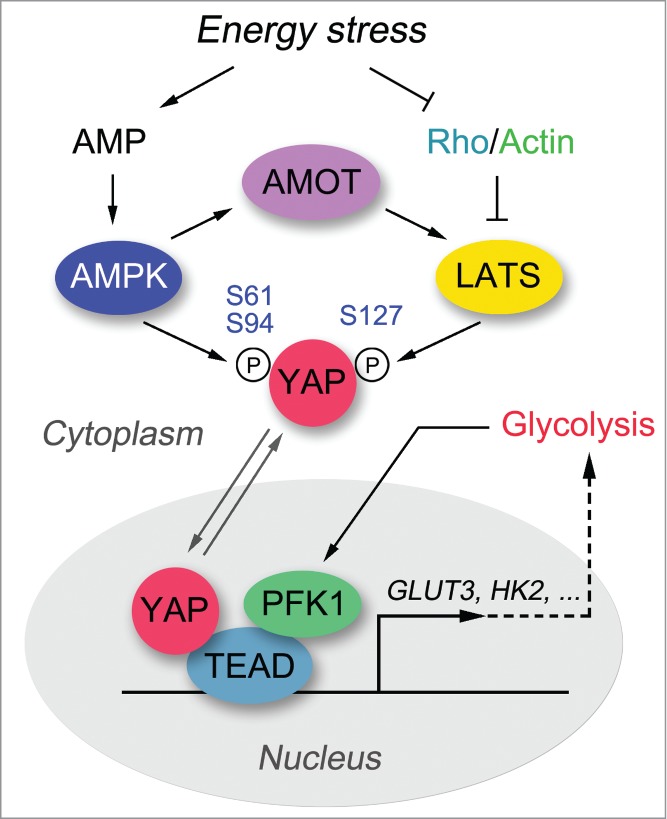
Energy stress regulates the Hippo-YAP pathway. Both AMPK and LATS kinases are activated under energy stress, and once activated phosphorylate YAP at several residues, contributing to the inhibition of YAP transactivation activity. Release from energy stress leads to translocation of YAP into the nucleus, where it forms a complex with TEAD to initiate downstream gene transcription. In addition, a glycolysis enzyme PFK1 has recently been shown to interact with TEAD and maintain the YAP-TEAD complex in the nucleus. The active YAP-TEAD complex promotes cell proliferation, anti-apoptosis, and glycolysis, in part by upregulating GLUT3 and possibly also HK2.
Although both AMPK and LATS are capable of phosphorylating YAP under energy stress, it is still not clear how they are coordinated in this process. Our study showed that AMPK functions independently of the Hippo pathway under energy stress.1 On the other hand, LATS kinase activation is partially regulated by AMPK,1,2 and this regulation may be mediated at least in part by the angiomotin (AMOT) family proteins.3 Besides AMPK, Rho GTPase also plays a critical role in transducing the energy stress signal to the LATS kinase.1 However, the upstream events that dictate Rho GTPase activity under energy stress remain unclear. Notably, we did not detect dramatic AMPK-mediated phosphorylation of TAZ, another LATS downstream effector in the Hippo pathway that shares sequence and functional similarities with YAP, indicating that TAZ and YAP may be regulated differently under energy stress. Of course, the interplay between the Hippo-YAP pathway and energy homeostasis is complex. For example, a recent study indicated that glycolysis enzyme phosphofructokinase (PFK1) could directly regulate the YAP-TEAD complex and promote its downstream gene transcription (Fig. 1), occurring independently of the LATS and AMPK kinases.4 Together, these studies provide several mechanisms for the energy stress—dependent regulation of the Hippo pathway.
Our studies also elucidate the oncogenic role of YAP in promoting glycolysis (Fig. 1),1 which is in part mediated by YAP-dependent upregulation of GLUT3. Consistent with this, the YAP downstream gene signature and the gene signature associated with glycolysis positively correlate in primary human breast cancers.4 These findings further suggest multi-level crosstalk between cellular metabolism and the Hippo-YAP pathway. Whether other key metabolic events besides glucose metabolism are regulated by YAP deserves further investigation.
YAP plays critical roles in promoting cell proliferation and anti-apoptosis, which indicates that cellular growth conditions should have an intimate relationship with YAP activation. Indeed, high cell density,5 serum deprivation6 and glucose starvation1,2 are all obstacles for cell growth, where YAP activities are highly suppressed. Interestingly, our unpublished data suggest that manipulation of cell growth conditions could affect YAP activation. For example, when we subjected confluent MCF10A cells to energy stress (i.e. glucose starvation or treatment with 2-DG), YAP localized dominantly in the cytoplasm under these 2 unfavorable growth conditions. However, YAP quickly translocated into the nucleus in these cells when energy stress was released, despite that these cells were still confluent. These observations suggest that YAP is highly alert to environmental cues and may integrate multiple signals to facilitate cell proliferation and survival.
Cancer cells are known to reprogram their metabolism and signaling pathways to support uncontrolled proliferation and survival during tumor progression; such alterations are considered hallmarks of cancer.7 Our findings on the connection between glucose metabolism and the Hippo pathway will not only enrich our understanding of these critical physiological events, but more importantly will provide new insights into potential cancer therapies that take advantage of these connections.
References
- 1.Wang W, et al.. Nat Cell Biol 2015; PMID:25751139; http://dx.doi.org/ 10.1038/ncb3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mo JS, et al.. Nat Cell Biol 2015; PMID:25751140; http://dx.doi.org/ 10.1038/ncb3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeRan M, et al.. Cell Rep 2014; 9:495-503; PMID:25373897; http://dx.doi.org/ 10.1016/j.celrep.2014.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enzo E, et al.. EMBO J 2015; pii: e201490379; PMID:2579644625796446 [Google Scholar]
- 5.Zhao B, et al.. Genes Dev 2007; 21:2747-61; PMID:17974916; http://dx.doi.org/ 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu FX, et al.. Cell 2012; 150:780-91; PMID:22863277; http://dx.doi.org/ 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns RA, et al.. Nat Rev Cancer 2011; 11:85-95; PMID:21258394; http://dx.doi.org/ 10.1038/nrc2981 [DOI] [PubMed] [Google Scholar]


