Figure 5.
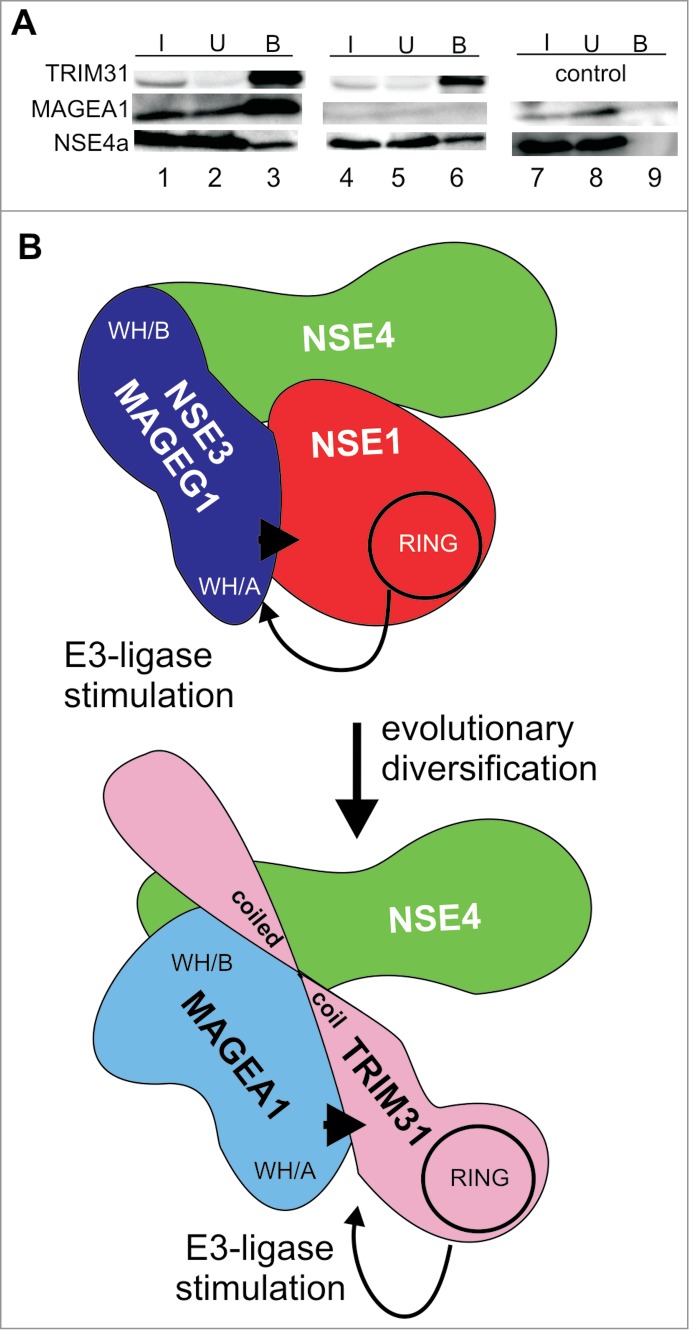
TRIM31 can form TRIM31-MAGEA1-NSE4 complexes reminiscent of NSE1-NSE3-NSE4 trimers. (A) myc-FLAG-TRIM31 (top panel, lanes 1–6), MAGEA1 (middle panel, lanes 1–3 and 7–9) and His-S-tag-NSE4a (bottom panel, lanes 1–9) constructs were co-transfected into HEK293T cells. The cell extracts were precipitated using the anti-FLAG-beads and immunoprecipitated proteins were analyzed on western blots. TRIM31 was detected using anti-myc, NSE4a was visualized using protein-S-HRP conjugate and the MAGEA1 protein was detected using anti-MAGEA1 antibody. Note that the endogenous levels of MAGEA1 in HEK293T cells are very low (lanes 4–6) compared to the levels of ectopically expressed MAGEA1 (lanes 1–3). These data suggest that TRIM31 binds directly to NSE4a and forms TRIM31-MAGEA1-NSE4a complex. (B) Cartoon comparing common features of the NSE1-NSE3-NSE4 and the TRIM31-MAGEA1-NSE4 complexes. The NSE3/MAGE proteins share conserved hydrophobic pockets (within the WH/B subdomain) interacting with NSE4/EID proteins. NSE3/MAGEG1 (WH/A) binds to NSE1 and stimulates (arrow) its RING-finger (RING) E3 ubiquitin-ligase activity. Similarly, MAGEA1 binds to TRIM31 and stimulates (arrow) its RING-finger (RING) E3 ubiquitin-ligase activity. Both, NSE3/MAGEG1 and MAGEA1, require their WH/A binding motifs for their stimulatory effects (arrowheads). As the TRIM31-MAGEA1-NSE4 complex shares common features with the yeast and human NSE1-NSE3-NSE4 complexes we propose that they may have evolved from a common ancestral NSE1-NSE3-NSE4 complex.
