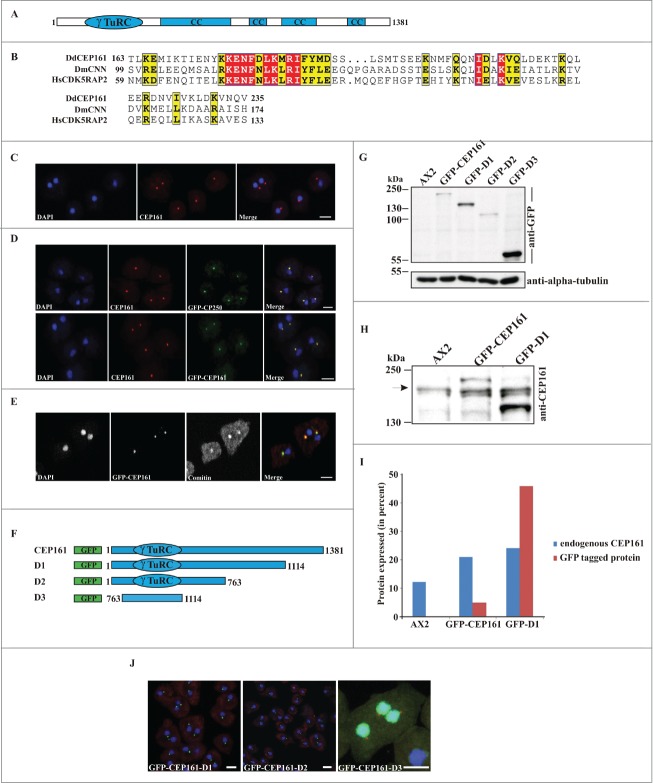
Figure 1.
(See previous page). CEP161 as a novel centrosomal protein in D. discoideum. (A) CEP161 protein and domain structure. (B) γ-TuRC domain sequence alignment. Protein accession numbers: Dictyostelium discoideum (Dd) (DDB_G0282851), Drosophila melanogaster (Dm) centrosomin (CNN) (NP_725298.1) and Human (Hs) (NP_060719). The numbers indicate the amino acid position of the γ-TuRC domain in the respective proteins. Color code: Red background, identical residues; yellow background, similar residues. (C) Localization of CEP161 at the centrosome in AX2 cells. CEP161 was detected with mAb K83-632-4. (D) Colocalization of CEP161 and GFP-CP250 at the centrosome (upper panel). mAb K83-632-4 recognizes GFP-CEP161 at the centrosome (lower panel). (E) Colocalization of GFP-CEP161 with comitin at the Golgi complex. Comitin was detected with mAb 190-340-8. DAPI was used to stain the nucleus. Scale bar, 10 μm. (F) Overview of N-terminally GFP-tagged CEP161 proteins. (G) Expression of GFP-tagged CEP161 proteins in AX2 cells. Proteins from whole cell lysates were separated by SDS-PAGE (10% acrylamide). mAb K3-184-2 was used to detect the GFP-fusion proteins Alpha-tubulin detected with mAb YL1/2 was used as the loading control. (H) Detection of endogenous CEP161 and GFP-tagged proteins with polyclonal antibodies specific for CEP161. The proteins were resolved by SDS-PAGE (10% acrylamide). The arrow indicates the position of endogenous CEP161. (I) Quantification of the protein gel blot results for amounts of endogenous CEP161 and GFP-tagged proteins. (J) Subcellular localization of GFP-tagged proteins. DAPI was used to stain the nucleus, the centrosome in D3 cells was stained with CP250 specific mAb K68-439-8. Scale bar, 5 μm.