Current approaches to seeking effective strategies of fighting cancer cells can be subdivided into 2 groups (1) improvement of the classical chemotherapeutics possessing general toxicity, and (2) the development of targeted therapies aimed at the elimination of cancer cells through destruction of their specific molecules or at the enhancement of immune response. The first strategy is limited by serious side effects, and the second does not always work because of high variability of the target determinants in cancer cells. An alternative strategy to overcome these limitations entails an impact on the RNA pool of cancer cells as a significant component of these cells that is different from normal cells. The degradation of RNA by exogenous RNases leading to death of cancer cells is a process that involves several stages. It includes interaction of RNase with the cell surface structures such as phospholipids, glycoproteins, and receptors, and with the endocytosis proteins clatrin, dynamin, and actin.1 Internalization of RNases is thought to be a cytotoxicity-limiting step. RNAs that are not associated with proteins, such as tRNAs and non-coding microRNAs (miRNAs), are most accessible for RNases. Especially important is our finding that the toxic effect of binase, the extracellular highly cationic RNase from Bacillus intermedius, toward cancer cells is due to the expression there of specific oncogenes e.g. KIT, ras, AML1/ETO and FLT3.1 We have shown also that RNase 5K Sa (cationic mutant of Streptomyces aureofaciens RNase) inhibits proliferation of ras-transformed fibroblasts, whereas this effect did not occur in normal cells and in cells transformed by fms and src oncogenes.2 Likewise, onconase (RNase from bullfrog Rana pipiens) exhibits cytotoxic activity toward ras-transformed mouse fibroblasts.3 Evidently, patterns of oncogene expression are linked to the selective cytotoxicity of exogenously applied RNases in cancer cells.
Recently there has been notable progress in research on antitumor RNases. The downstream mechanisms of RNase cytotoxicity may induce regulatory activity of products of RNA hydrolysis, as well as selective suppression of certain genes. RNases have the potential to destroy RNA in cancer cells and produce the generation of damage-associated molecular patterns, which can stimulate immune sensors such as toll-like receptors. Once activated, they initiate signaling pathways leading to the release of cytokines and chemokines, which recruit immune cells inducing further cytokine production, the production of angiogenic mediators and growth factors, all of which may influence tumor progression.4
The question is what kind of RNases will be effective in fighting cancer? First of all, they must have an ability to effectively interact with the cell surface. Net positive charge of RNase molecule contributes to this ability. Tumor cells express much more acid phospholipids and glycoproteins in the membrane outer leaflet than their non-tumor counterparts; therefore, cytotoxic RNases have to be cationic molecules. Indeed, all of the known RNases possessing antitumor activity are cationic proteins: onconase, bovine seminal RNase, eosinophil cationic protein, binase, and B. amyloliquefaciens RNase barnase.1 Two isozymes, RNases Sa and RNase Sa3, differ in the net charge (pI = 3.5 and 5.4, respectively), and a more cationic RNase is much more cytotoxic.5 Recently, we have constructed a set of RNase Sa variants consisting of charge reversal mutants, charge neutralization mutants, and variants with positively charged cluster at the N-terminus. We have tested all variants on acute myeloid leukemia cells Kasumi-1 and shown that cytotoxicity of the catalytically active RNase Sa mutants is linearly enhanced by cationization, and that cells die by apoptosis.6 Moreover, the net charge of RNase molecule seems to be more important for antitumor effects than its distribution: localization of positive charge on the N-terminus does not contribute to RNase Sa cytotoxicity.
In Figure 1 we have schematically represented the stages of exogenous RNases interaction with tumor cell, leading to apoptotic death of the latter. The RNase capability of inducing apoptotic cell death provides an important argument for the potential use of RNases to eliminate tumors. After internalization, cytotoxic RNases activate the program of apoptotic cell death. Binase was the first bacterial RNase that demonstrated noticeable apoptogenic activity against cancer cells.5 In particular, we have shown that binase-induced tumor cell death has signs of both mitochondrial and ligand-dependent apoptosis.7 Importantly, the effect of binase does not lead to necrotic processes in cell cultures as well as in vivo for the entire organism (murine tumor model).7 At the same time binase effectively inhibits primary tumor growth and metastases in animals with induced tumors. Moreover, binase triggers regenerative processes in the liver of mice with Lewis lung carcinoma and has no general toxic effect on the organism in normal mice.7 We believe that in the near future new experimental data will be obtained, ensuring a deeper insight into the molecular mechanism of selective toxic effect of cationic RNase on malignant cells, and that these enzymes will serve as valuable contribution to the fight against cancer.
Figure 1.
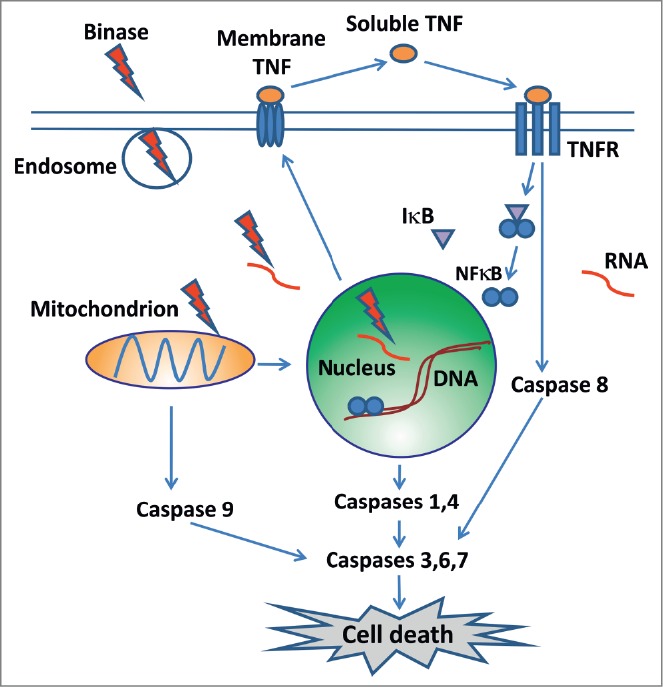
Binase penetrates into the cells in the endosome; attacks a target RNA; affects gene expression; promotes apoptosis by activating the mitochondrial pathway through caspase 9. Binase activates TNF, which mediates apoptosis through caspase 8, and also activates NFκB. NFκB in nucleus promotes the expression of caspases 1 and 4. Activation of caspases 3,6 and 7 leads to cell death.
References
- 1. Mitkevich VA, et al. Mol Biol 2014; 48:181-8; http://dx.doi.org/ 10.1134/S0026893314020137 [DOI] [Google Scholar]
- 2. Ilinskaya ON, et al. Protein Sci 2002; 11:2522-5; PMID:12237473; http://dx.doi.org/ 10.1110/ps.0216702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smith M, et al. Exp Cel Res 1999; 247:220-32; PMID:10047464; http://dx.doi.org/ 10.1006/excr.1998.4317 [DOI] [PubMed] [Google Scholar]
- 4. Rich A, et al. Front Immunol 2014; 24:464; PMID:25309546; http://dx.doi.org/ 10.3389/fimmu.2014.00464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sevcik J, et al. J Biol Chem 2002; 277:47326-30; PMID: 12228255; http://dx.doi.org/ 10.1074/jbc.M208425200 [DOI] [PubMed] [Google Scholar]
- 6. Mitkevich VA, et al. Oncoscience 2014; 1:738-44; PMID:25594000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mironova NL, et al. Cell Cycle 2013; 12:2020-31; PMID:23759588; http://dx.doi.org/ 10.4161/cc.25164 [DOI] [PMC free article] [PubMed] [Google Scholar]


