Figure 4.
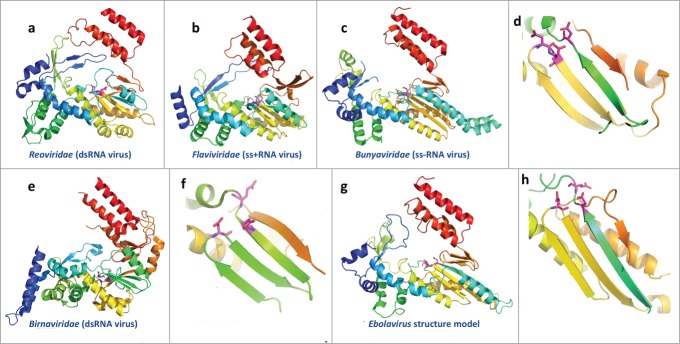
Structures of the catalytic domains of RNA-dependent RNA polymerases (RdRP) from RNA viruses and the structure model for Ebolavirus RdRP. The virus family is labeled below. The structures are colored in rainbow, with equivalent secondary structure elements from different structures colored similarly, except for the Birnaviridae RdRP, which has a circularly permutated topology. The functional sites used to coordinate Mg2+ are shown as sticks and colored in magenta. (A–C) Overall structure of the core domains of RdRPs from RNA viruses (PDB IDs: 2R7O, 1GX5, and 5AMR); (D) close up view of the classic arrangement of functional sites for the core domains of RdRPs from RNA viruses; (E–F) overall structure and close up view of the functional sites for the RdRP from Birnaviridae (dsRNA virus); PDB id: 2PGG; (G–H) structure model for the core domains of ZEBOV RdRP and close up view of the predicted active sites.
