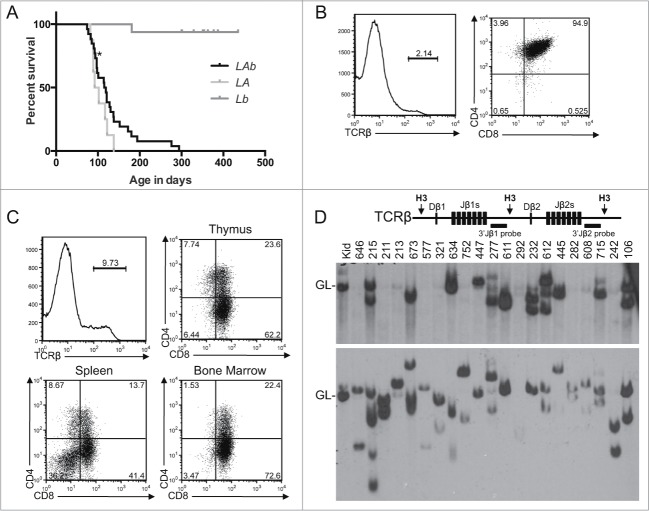
Figure 1.
T-ALL Predisposition of Atm−/− Mice with Pervasive Mono-Allelic Deletion of Bcl11b initiating in DN Thymocytes. (A) Kaplan-Meier curve depicting the cancer-free survival of parallel cohorts of 26 LAb, 8 LA, and 16 Lb mice. All cohort mice succumbed to thymic T-ALLs except for LAb mouse #211 that succumbed to a T-ALL in peripheral lymphoid tissues (indicated by asterisk). These mice were of a mixed C57BL6 and 129SvEv background.(B-C) Flow cytometry analysis of LAb T-ALLs no. 321 (B) and no. 277 (C) showing surface expression of TCRβ or CD4 and CD8. This analysis was conducted as described (33). Gates were drawn using normal thymocytes or splenocytes. The percentages of cells in each gate are indicated. (D) Schematic of the Tcrβ locus (top) and Southern blot analyses of Tcrβ rearrangements (bottom). Top, shown are relative locations of the indicated Tcrβ segments, HindIII restriction sites (H3), and 3′Jβ1 and 3′Jβ2 probes. Bottom, Southern blots of HindIII-digested DNA from the indicated LAb thymic cancers (and LAb splenic cancer #211) or from the kidney of a WT mouse using the 3′Jβ1 or 3′Jβ2 probe as previously described.33 Germline (GL) bands for each probe are indicated. The membrane was hybridized with the 3′Jβ1 probe and then stripped and hybridized with the 3′Jβ2 probe, revealing which LAb cancers lack 3′Jβ1-hybridizing band(s) due to Vβ-to-Dβ2-Jβ2 rearrangements on both alleles. The images were cropped from a larger blot. Mouse no. 634 was removed from the cohort for incorrect genotyping.