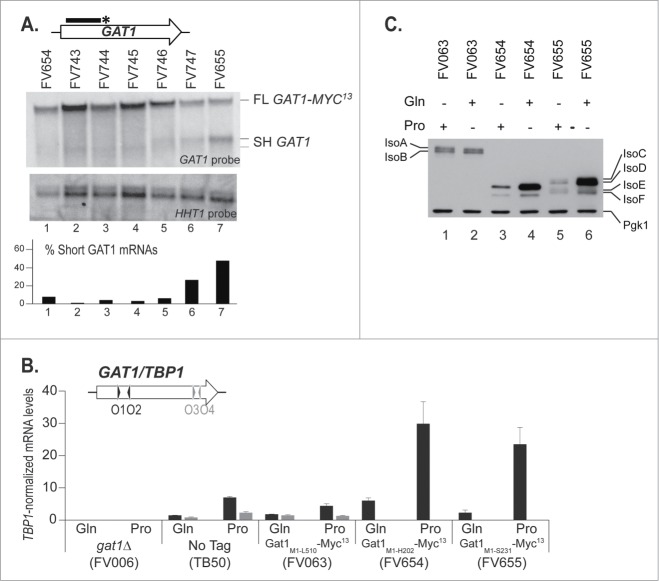
Figure 5.
C-terminal deletions to identify the site for premature transcription termination at the GAT1 locus. gat1Δ (FV023), wild type untagged (TB50), GAT1-MYC13 (FV063), GAT11–606-MYC13 (FV654), GAT11–621-MYC13 (FV743), GAT11–636-MYC13 (FV744), GAT11–651-MYC13 (FV745), GAT11–666-MYC13 (FV746), GAT11–681-MYC13 (FV747) and GAT11–693-MYC13 (FV655) cells were grown in YNB medium with glutamine (Gln) or proline (Pro) as the nitrogen source. (A). Total RNA was isolated and 30 μg from each sample were subjected to Northern blot analysis using a double stranded GAT1-specific probe covering the 5′ region of the gene. HHT1 was used as the loading and transfer efficiency control. Myc13-tagged mRNAs and prematurely terminated transcripts were quantified. Histograms in the lower part of the panel indicate the proportion of prematurely terminated transcript versus total GAT1 mRNAs within each experiment. (B). Total RNA was isolated and GAT1 mRNA levels were assessed using qRTPCR as described in Figure 3 using GAT1O1-O2 and GAT1O3-O4 primer pairs. (C). Gat1 protein species were analyzed with anti-myc western blotting as described in material and methods. Loading uniformity was assessed using anti-pgk1 antibodies.