Figure 1.
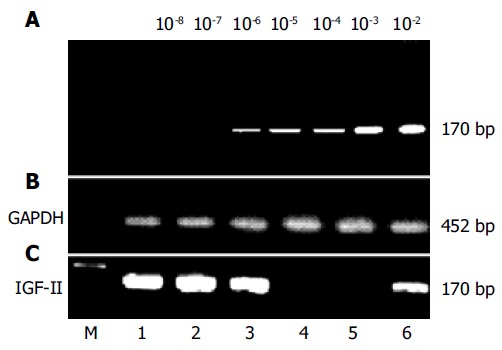
Amplification of IGF-II genomes from human hepatoma tissues or circulating blood samples of patients with hepatocellular carcinoma. IGF-II mRNAs were synthesized according to IGF-II cDNA with random hexamers and moloney murine leukemia virus reverse-transcriptase, and detected with different primer pairs by nested PCR (170 bp). The positive fragments of IGF-II genome were found distinctly in hepatoma tissues or in peripheral blood of patients with hepatocellular carcinoma. A: the sensitive limitation of our detection system (2 ng/L), using total RNA with 10-2-10-8 fold dilution and then amplified by nested PCR; B: the amplified fragments (452 bp) of glyceraldehyde-3-phosphate dehydrogenase genome from liver tissues or peripheral blood as controls; C: the amplification of IGF-II genomes in liver tissues (No. 1-4) or circulating blood (No. 5-6). No. 1-2, the positively amplified fragments of IGF-II mRNA from cancerous tissues of patients with hepatocellular carcinoma; No. 3, the positively amplified fragments of IGF-II mRNA from para-cancerous tissue of patients with hepatocellular carcinoma; No. 4, no positively amplified fragment from non-cancerous tissue of patients with hepatocellular carcinoma; No. 5, no positively amplified fragment from circulating blood of patients with liver cirrhosis, and No. 6, the positively amplified fragment from peripheral blood of patients with hepatocellular carcinoma. GAPDH: glyceraldehyde-3-phosphate dehydrogenase. M: DNA molecular weight marker.
