Figure 2.
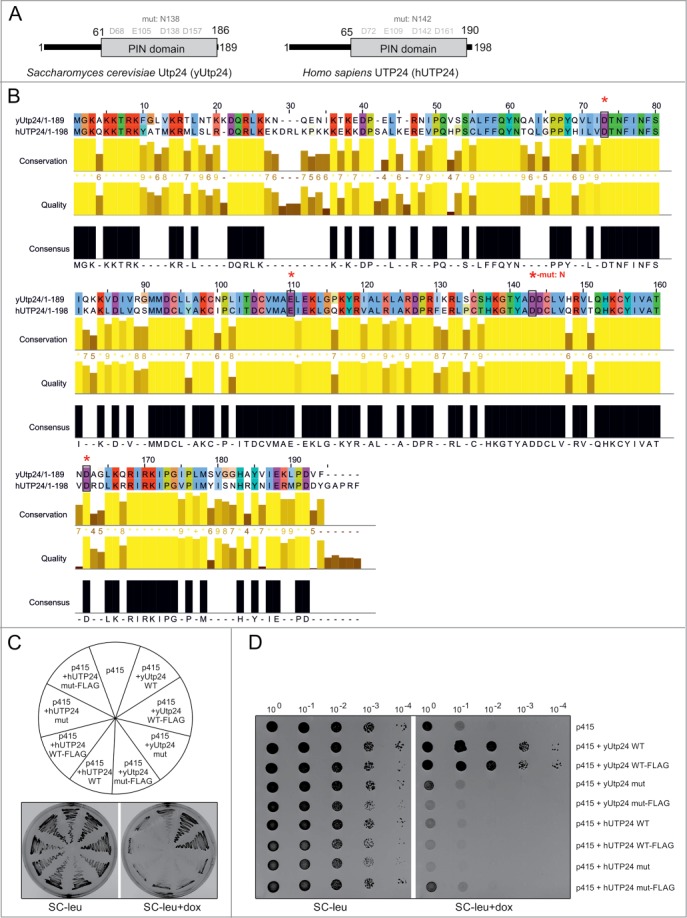
hUTP24 does not complement yUtp24 depletion in S. cerevisiae. (A) Schematic view of yeast and human UTP24 proteins. Both proteins are less than 200 amino acids in length, with an unstructured region at the N-terminus. This is followed by the PIN domain, which is potentially associated with ribonuclease activity. Conserved D-E-D-D tetrads of acidic residues in the putative active site of the PIN domain are highlighted in gray based on the detailed alignment in (B). Positions of D to N substitutions, introduced into yUtp24 (amino acid 138) and hUTP24 (amino acid 142), to generate mutant variants of the proteins are indicated above. (B) Detailed amino acid alignment of yUtp24 and hUTP24. Evolutionarily conserved, negatively charged residues are marked with black rectangles and red asterisks. The position of a D to N amino acid change in the mutant versions of either protein is indicated. (C and D) hUTP24 is not able to replace yUtp24 function in yeast. An S. cerevisiae strain in which endogenous yUTP24 had been placed under the control of a doxycycline-repressible promoter was transformed with an empty p415 vector or its derivatives encoding WT and mut variants of yUtp24 or hUTP24 that possess or lack a FLAG-tag at the C-terminus. In (C), growth of the transformants was analyzed by streaking approximately equal amounts onto media with or without doxycycline. In (D), 10-fold serial dilutions of the liquid cultures of the respective transformed strains were analyzed for growth in the absence or presence of doxycycline. In both cases, plates were incubated for 48 hours at 30°C.
