Figure 3.
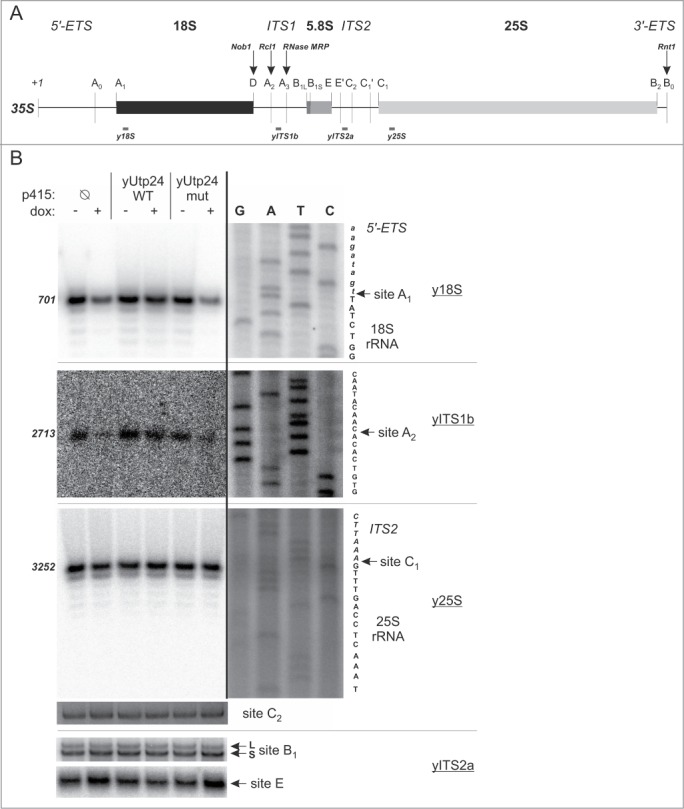
Mutation in the putative active site of yUtp24 leads to inefficient processing of pre-rRNA at sites A1 and A2. (A) A scheme of the S. cerevisiae 35S pre-rRNA. Positions of the processing sites within the 35S precursor are indicated with vertical thin lines together with the names. Nucleotide numbering is based on the GenBank 35S pre-rRNA sequence (accession number: BK006945.2). Arrows indicate sites of endoribonucleolytic processing carried out by known enzymes – their names are provided above with bolded italics. Gray bars below the primary 35S transcript show positions of oligonucleotides used for primer extension in this study. (B) Identification of processing defects by primer extension. The S. cerevisiae strain with endogenous yUTP24 under the control of a doxycycline-regulated promoter was transformed with an empty p415 vector (Ø) or its derivatives encoding WT and mut variants of yUtp24 or hUTP24. Total RNA was isolated from transformants grown either in the absence (“dox: −”) or in the presence (“dox: +”) of doxycycline and subjected to primer extension analysis using oligonucleotides (marked by underline) hybridizing upstream selected processing sites in 35S pre-rRNA (indicated by arrows). Numbers on the left indicate nucleotide positions in 35S corresponding to reverse transcription stops. The same primers were used in parallel with DNA templates comprising appropriate rDNA fragments to generate dideoxynucleotide sequencing ladders, which were co-electrophoresed with primer extension products. Retrieved sequences (in reverse complement) are presented next to the sequencing results. Note that various exposures of gel fragments are presented to optimally visualize the data obtained.
