Figure 4.
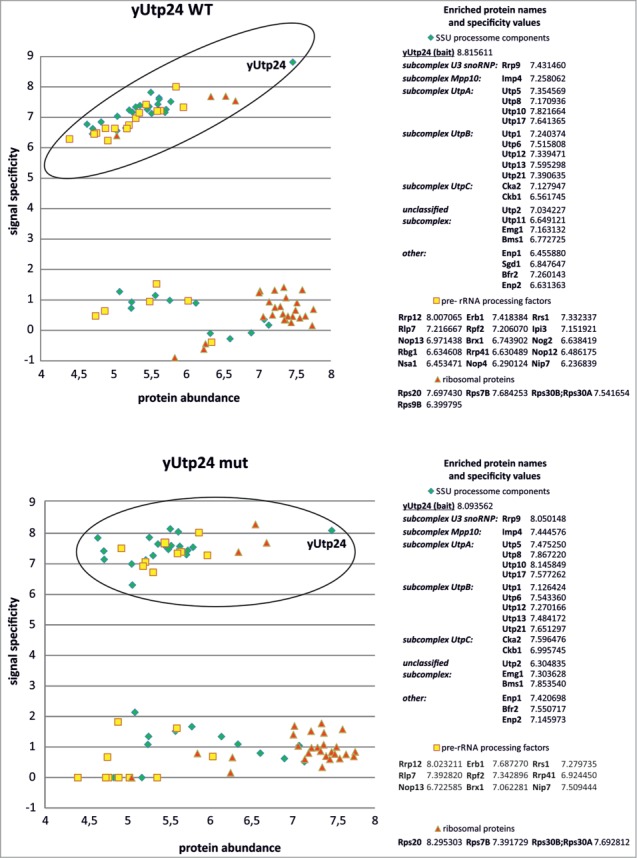
yUtp24 protein interacts with SSU processome components as well as with other factors involved in yeast ribosome biogenesis. Wild-type (top) and mutant yUtp24 (bottom) were purified from the yeast strains as C-terminal fusions with a TAP-tag. Proteins co-purifying with the bait were analyzed by mass spectrometry and the results were compared to the parallel purification, carried out using the unmodified parental strain. To determine whether a given protein specifically co-purified with yUtp24, we compared its intensity to the intensity of that protein in the control sample. Signal specificity (y-axis) was defined as the log10 of the ratio of protein signal intensity measured in the bait purification to background level (which is the protein signal intensity in the negative control purification; background level was arbitrarily set to 1 for proteins not detected in the negative control). Protein abundance (x-axis) was defined as the log10 of the ratio of protein signal intensity divided by its molecular weight in kDa. This parameter was implemented to eliminate differences due to the size of proteins. Points located within the ellipses correspond to high values of both protein abundance and specificity, indicating proteins enriched in bait purification (compared to the control sample) and thus suggest interaction. These hits (subdivided into 3 categories: SSU processome components, pre-rRNA processing factors, and ribosomal proteins) are listed next to the graphs and the calculated specificity values are indicated. In the case of SSU processome subunits, identified proteins belonging to specific subcomplexes within the assembly are indicated, according to ref. 50. The remaining points represent proteins that are present in similar amounts in both bait purification and the control sample.
