Figure 3. miR-21 completely disrupted clones derived from clone #38 using TALEN L2-R2.
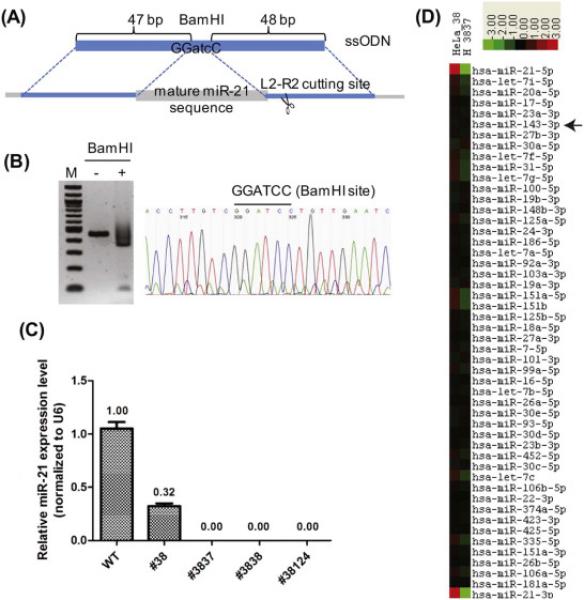
(A) A homologous recombination event between the single-stranded oligodeoxynucleotides (ssODN) and the miR-21 genomic locus will delete the miR-21 sequence and replace with 3 bases of atc to generate a BamHI site. (B) Genomic DNA from clone #38124 was subjected to PCR and BamHI digestion (left) and Sanger sequencing (right). (C) Mature miR-21 levels were measured by RT-qPCR. (D) A heatmap of the miR-Seq data from clones #38 and #3837. The 49 most abundant microRNAs plus miR-21-3p were ranked and clustered by centering on the row-wise mean log(2) value [15, 17]. A scale of 8-fold deviation from the mean is shown.
