Figure 3.
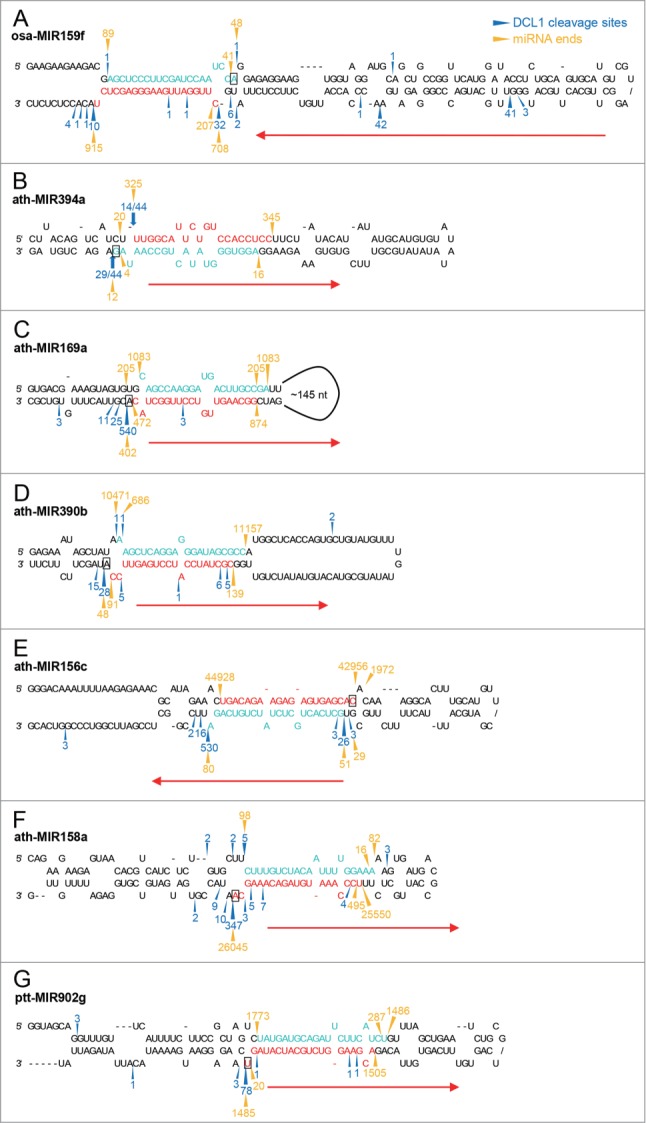
Comparison of miRNA termini with DCL1 cleavage sites. (A) Two major DCL1 cleavage sites revealed by rice PARE data define 20-nt osa-miR159f. The positions and counts of DCL1 cleavage on osa-MIR159f were based on rice PARE data published by Li et al.44 (B) Cycle RT-PCR reveals DCL1 cleavage sites consistent to the 5′ end of 20-nt miR394 and the 3′ end of 21-nt miR394* which form a 3-nt overhang. (C and D) DCL1 cleavage sites are mapped to the position 1 nt downstream of 20-nt ath-miR169a* (C) and ath-miR390b* (D) 3′ ends. The positions and counts of DCL1 cleavage on ath-MIR169a and ath-MIR390b were based on A. thaliana SPARE data published by Bologna et al.13 (E–G) Major DCL1 cleavage sites are consistent to the 5′ and 3′ termini of 20-nt miRNA duplexes with an 18-bp duplex region. The positions and counts of DCL1 cleavage on ath-MIR156c (E), ath-MIR158a (F), and ppt-MIR902g (G) were based on A. thaliana SPARE data and P. patens degradome data published by Bologna et al. and Addo-Quaye et al., respectively.13,42 Sequences of 20-nt miRNAs and their complementary strands identified with small RNA data are highlighted in red and turquoise, respectively. Blue arrowheads and numbers indicate DCL1 cleavage sites and counts based on degradome or SPARE data. Blue arrows and numbers indicate the positions and frequency of DCL1 processing sites detected by cycle RT-PCR. Gold arrowheads and numbers indicate the termini and counts of the 2 most abundant small RNA sequences overlapping with the annotated miRNAs for each strand. Red arrows indicate the directions of DCL1 processing and the nucleotides presumably associated with the putative 3′ binding pocket of DCL1 are indicated with rectangles.
