Figure 2.
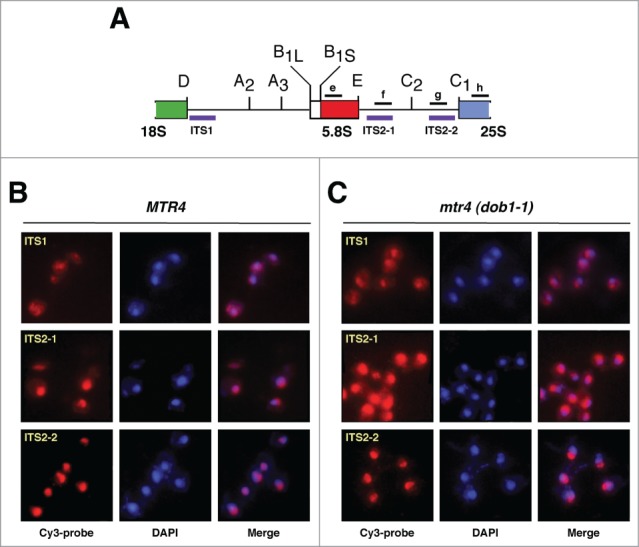
Detection of pre-ribosomal particles by fluorescence in situ hybridization (FISH). (A) Primary structure of the 2 internal transcribed spacers, ITS1 and ITS2, of the 35S pre-rRNA. Processing sites are shown. The location of the 3 Cy-3 labeled probes used for FISH (purple) and of the oligonucleotides probes used for northern hybridization (black) is indicated. (B and C) Wild-type (MTR4) and dob1–1 (mtr4) cells were grown at 30°C in YPD medium to mid-log phase. Distinct pre-rRNAs were detected by FISH using Cy-3 labeled ITS1, ITS2–1 or ITS2–2 probes (red; purple in merge panels). Cells were further stained with DAPI to visualize the nucleoplasm (blue). All images shown were captured using identical exposure times and processed in the same manner.
