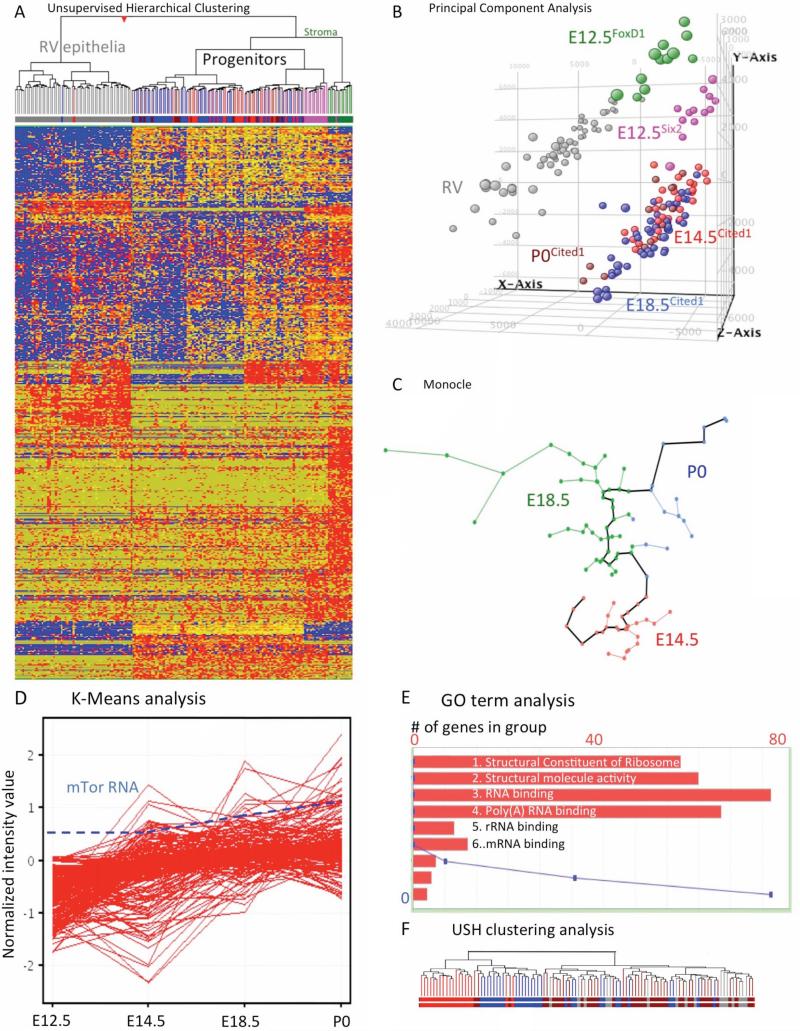
Figure 7. Singe cell RNA-seq analysis of Cited1+ progenitors.
(A) A heat map depicting the expression of genes in Cited1+ progenitors (E12.5, E14.5, E18.5 and P0), stromal cells (FoxD1+, E12.5) and RV cells. Unsupervised clustering visualize the expression patterns of genes (E12.5 magenta; E14.5 red; E18.5 blue; P0 brown; Foxd1+ stroma green; RV grey). Red, blue and yellow intensities indicate high, low and intermediate expression levels, respectively. (B) Analysis of single-cell RNA-Seq data using principle component analysis (PCA) (C) The algorithm Monocle was utilized to organize the cells using “pseudo-time analysis” of E14.5, E18.5 and P0 Cited1+ cells. (D) Gene expression profile from K-means cluster #1 showing the increase of ribosomal biosynthesis genes during progenitors aging. A dashed line represents the concomitant increase in expression of mTOR mRNA. (E) Graphic illustration showing the top five GO terms from K-means cluster 1. The X-axis displays the number of genes in the specified GO term, the Y-axis plots the p-value as a blue line. (F) Hierarchical clustering of cells based on genes in K-means cluster #1 shows the segregation of progenitors into three major clusters: one containing only E12.5 progenitors, a cluster containing E12.5, E14.5 and a few E18.5 progenitors and a large cluster containing only E18.5 and P0 progenitors. See also Table S2-7.