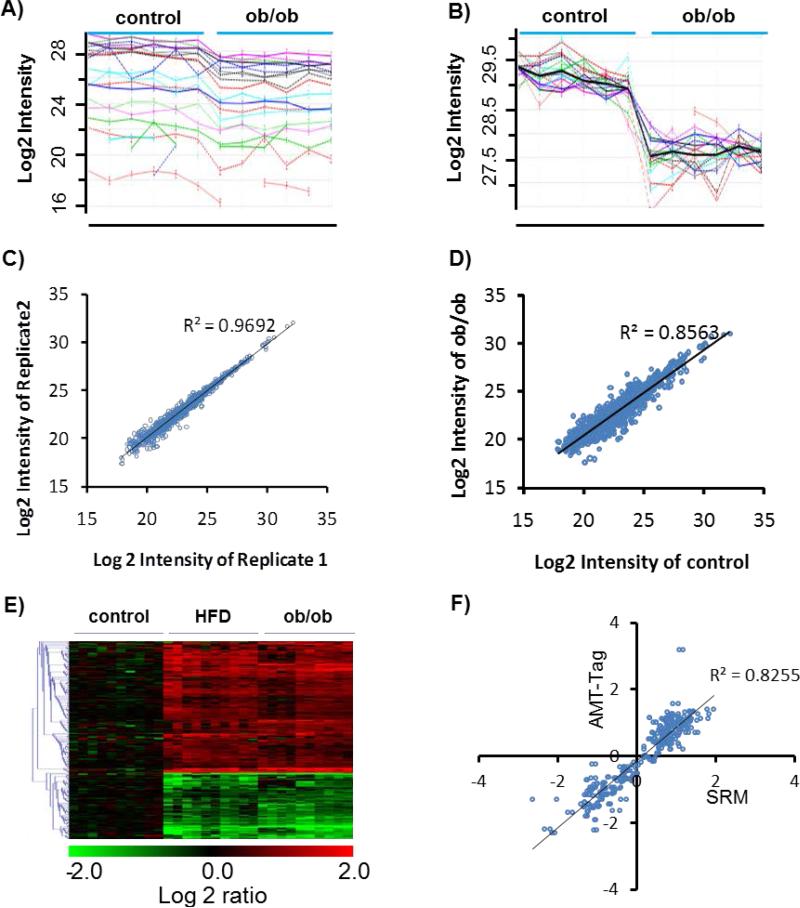
Fig.2. Quantitative analysis strategy for the islet proteome.
A, Raw peptide intensity profile data (log2 transformed) of all peptides identified from glucagon. Each line profile represents a peptide from glucagon. 3 Control and 3 ob/ob (small islets) samples are presented with each sample analyzed in duplicated. B, Rescaled peptide intensity profile data. Dark line represents the protein abundance profile by averaging the intensity of all peptides after the rescaling process. C, Reproducibility of protein abundance quantification between technical replicates. D, Comparison of protein abundances between control and ob/ob (small islets). E, Heatmap of all proteins with significant changes. Each condition has 5 biological replicates, each replicate has duplicated runs. Values are normalized to the average of control. Only data from small islets from ob/ob mice were presented here. F, Validation of label-free quantitative data for selected proteins by selected reaction monitoring (SRM). Values are the log2 ratios to control. Each data represents one protein in one condition summarized from 5 biological replicates. Data points from both HFD and ob/ob mice were included here.