Figure 1. Chemical proteomic probes for mapping lipid-binding proteins in cells.
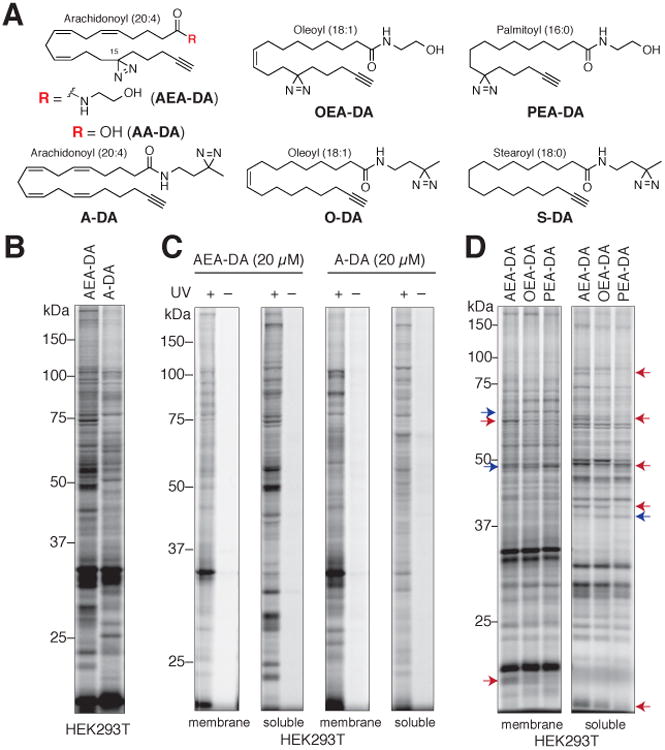
(A) Structures of lipid probes featuring arachidonoyl (AEA-DA, AA-DA and A-DA), oleoyl (OEA-DA and O-DA), palmitoyl (PEA-DA) and stearoyl (S-DA) acyl chains, as well as photoreactive (diazirine) and alkyne groups.
(B) AEA-DA and A-DA probes show overlapping, but distinct protein interaction profiles in HEK293T cells. Cells were treated with each probe (20 μM) for 30 min in situ before photocrosslinking and analysis of probe-modified proteins as described in Figure S1.
(C) Arachidonoyl probe labeling of membrane and soluble proteins depend on UV irradiation of cells.
(D) Comparative labeling profiles of lipid probes (20 μM, 30 min) in HEK293T cells. Red and blue arrows mark representative proteins preferentially labeled by arachidonoyl and oleoyl/palmitoyl probes, respectively. See Figure S1C for profiles of A-DA, O-DA and S-DA.
