Figure 7. Target engagement and lipid metabolism effects of NUCB1 ligands.
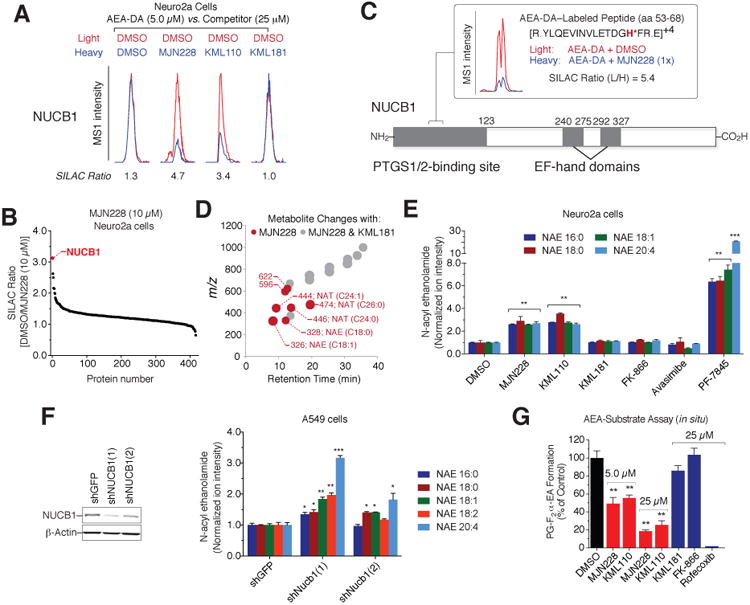
(A) Representative peptide MS1 chromatograms showing blockade of AEA-DA probe labeling of endogenous NUCB1 in Neuro2a cells by MJN228 and KML110, but not KML181.
(B) SILAC ratio plot for in situ competition experiment performed with MJN228 (10 μM) and the AEA-DA probe (5 μM).
(C) LC-MS/MS identification of a prominent MJN228-sensitive, AEA-DA-modified NUCB1 peptide (aa 53-68) in Neuro2a cells.
(D) Untargeted metabolite profiling reveals that Neuro2a cells treated with MJN228 (10 μM) show elevated fatty acid amides (NAEs and NATs) compared to cells treated with DMSO or KML181 (10 μM) (P < 0.0001, n = 5 per condition). See also Table S5.
(E) Targeted MRM measurements showing elevations in NAEs in Neuro2a cells treated withNUCB1 ligands MJN228 and KML110 (10 μM, 6 h), but not KML181, FK-866, or avasimibe. See Figure S7G for MRM measurements of NATs.
(F) Left, Western blot showing knockdown of NUCB1 in shNUCB1 A549 cell lines compared to a control cell line (shGFP). Right, both shNUCB1 cells show significant elevations in NAEs compared to the control shGFP cell line.
(G) NUCB1 ligands MJN228 and KML110 (5 and 25 μM), but not KML181 or FK-866 (25 μMeach), suppress the conversion of exogenous AEA (20 μM, 30 min) to PGF2α-EA in PMA-stimulated A549 cells. Rofecoxib (25 μM) also blocked PGF2 α-EA synthesis.
For (E-G), data represent mean values ± SEM; n = 3-4/group. *P < 0.05, **P < 0.01, ***P < 0.001 for DMSO-treated (E) or shGFP cells (F) versus compound-treated (E) or shNUCB1 (F) cells.
