Abstract
Life on Earth is dictated by circadian changes in the environment, caused by the planet's rotation around its own axis. All forms of life have evolved clock systems to adapt their physiology to the daily variations in geophysical parameters. The intestinal microbiome serves as a signaling hub in the communication between the host and its environment. We recently discovered that the microbiota undergoes diurnal oscillations in composition and function, and that these oscillations are required for metabolic homeostasis of the host. Here, we highlight these findings from the perspectives of microbial system stability and meta-organismal metabolic health. We also discuss the contribution of nutrition and biotic interventions on diurnal processes of the microbiota and their potential involvement in diseases commonly associated with circadian disruption.
Keywords: diurnal oscillations, food, metabolic disease, microbiota
Introduction: Circadian Rhythms as a Principle of Life on Earth
The temporal instability of the environment is an inherent property of life on Earth. Light, temperature, and availability of nutrients fluctuate on the scale of seasons, months, and days. These characteristics are met by the development of clock systems in all domains of life which couple organismal activity to the geophysical time, thereby synchronizing physiological processes to daily variations in environmental conditions.1
In mammals, this synchronization is achieved by a molecular clock comprising a network of several transcriptional factors which is present in virtually all cells of the body. Several features make mammalian clocks suitable to adapt to the conditions imposed by the daily geophysical fluctuations: (1) Circadian clocks are self-sustained and intrinsic; and (2) their rhythm can be entrained by environmental signals, called “Zeitgeber” (timing cue), including light, temperature, and feeding times.2 The coordination of circadian clocks within the body and across organs follows a hierarchical principle. The master pacemaker, located in the brain, is light-entrainable and influences the activity of peripheral clocks.3 Nonetheless, peripheral clocks can be dominantly entrained by food and thereby uncoupled from the activity of the master clock.4,5 On a molecular level, the circadian clock is driven by rhythmic interlocked negative feedback loop between the transcription factors BMAL1/CLOCK and Period/Cryptochrome. Additional feedback loops extend to the nuclear receptors of the ROR and REV-ERB families. This network coordinates the rhythmic expression of up to 15% of the entire transcriptome of each cell in a circadian manner.6
The prokaryotic circadian clock has primarily been studied in light-responsive cyanobacteria.7 Remarkably, in this system, a circadian clock of just 3 proteins (KaiA, KaiB, and KaiC) can be sustained even in the absence of transcription,8 although a transcriptional feedback is required for its stability.9 This suggests that molecular clocks can assume diverse forms and compositions across the domains of life. This is particularly interesting in the context of multi-domain ecosystems, in which the activity of eukaryotic and prokaryotic symbionts must not only be adjusted on one another, but also to the environmental fluctuations over the course of a day. It has recently been established that the community of intestinal bacteria colonizing a mammalian host, termed intestinal microbiome, constitutes such an ecosystem.
Temporal Stability of the Intestinal Microbiome
There is a growing body of evidence indicating significant contribution of the gut microbiome to host health and disease. The microbiome was shown to be an important effector of host metabolism, in energy harvest from food,10 changing host propensity toward weight-gain11,12 driving host metabolic stability13 or instability,14 and determining personal responses to biochemical compounds such as drugs15 and dietary supplements.16 The microbiome also works in coordination with the host immune system and can promote resistance to infection. A disrupted microbiome may exacerbate immune disorders such as colitis,17 and a healthy one can be used therapeutically to cure life-risking infections.18 The effect of the microbiome is not limited to the gut, and it may exert its influence systematically. One such recent example is featured in a mouse model of autism spectrum disorder, in which a healthy microbiome was shown to be able to ameliorate some neurological features of this disease.19 The extent of effect of the microbiome on all these aspects is governed by its composition and function, but in contrast to host genetics, which remain constant throughout its life, the composition and function of the gut microbiome is dynamic and potentially amenable to change.
Different stages of life are characterized by different microbiota configurations, which are assumed to have coevolved with host developmental stages.20 The initial inoculation of mammalians with gut microbes takes place at birth, when passing through the birth canal, and neonatal microbiomes are shaped by maternal microbes and maternal lactic compounds. These natural processes were shown to increase heterogeneity and diversity of the infant microbiome21 and to enhance immune specificity.22,23 Upon stabilization of the adult microbiome, its composition remains relatively constant, with a retention rate of approximately 60% over 5 years.24 However subtly, the composition of the microbiome maintains a rate of change well into old age, with distinct microbiomes in elderly subjects.25,26
While long-term compositional changes are important to the general understanding of host-microbiome coevolution and to the composition of the microbiome in health and disease, they only partially describe the variation landscape of the microbiome. The long-term resistance to perturbation may be overcome by diet, drugs and food supplements, in a process that depends both on the composition of the microbiome and on the type of perturbation. It is widely agreed that dietary changes are major drivers of microbiome variation. Restriction of caloric intake for a period of a year can cause a major and persistent shift in microbial composition, which is reflected primarily in the well-studied Firmicutes-to-Bacteroidetes ratio.27 Dietary perturbation may also cause more rapid and transient changes in microbial composition and function. A drastic change in fiber consumption was shown to shift microbial composition within 10 days, with a major part of the change showing 24 hours from perturbation.28 Another study has shown that switching to strictly plant-based or animal-based diets drives an enrichment of microbial functionality associated with the decomposition of plant-material or animal-material, respectively.29
Diurnal Oscillations of the Intestinal Microbiome
We recently revealed an additional layer of temporal fluctuations in the composition and function of the intestinal microbiome, which occurs at the scale of hours and follows diurnal rhythmicity.30 We found that up to 20% of all commensal species in mice and humans undergo diurnal fluctuations in their relative abundance, resulting in rhythmic changes of the entire bacterial community over the period of one day. This leads to time of the day-specific abundances of major components of the intestinal microbiota. For instance, the common mouse and human commensal genus Lactobacillus increases in relative abundance during the resting phase (the light phase in a mouse) and declines during the active phase.30 Of note, a recent study confirmed these observations, indicating that microbiota oscillations exist across animal facilities and housing conditions.31 Indeed, diurnal fluctuations in bacterial abundances are not tied to a particular microbiota configuration, but can be observed in mice and humans that feature inter-individual variability in the composition of commensal bacteria.30 Importantly, these fluctuations result in time of the day-specific functional profiles of the microbiome: energy harvest, DNA repair, and cell growth are primarily performed during the active phase of the host, while detoxification and chemotaxis are more abundant during the resting phase of the host (Fig. 1). This suggests that specialist members of the microbiota gain abundance at the expense of other commensals, or the entire biome content changes dynamically in response to food intake and nutrient availability, which results in temporal partitioning of the metabolic activity performed by the whole community.
Figure 1.
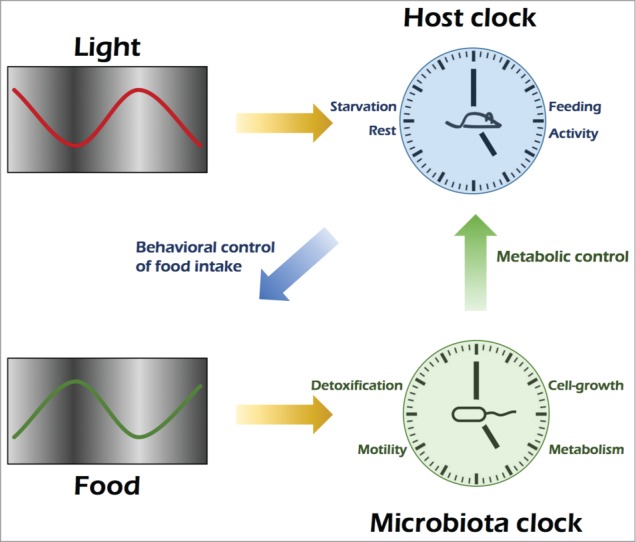
Model of cooperative host-microbial diurnality Light entrains the master clock of the host, whose rhythmic activity determines the time of food intake. Feeding times are controlling diurnal activity of the microbiome, which in turn is necessary for metabolic homeostasis of the host.
Interestingly, these temporal oscillations of the microbiome do not seem to be intrinsic and self-sustained, but require a functional clock of the host. Mice lacking PER1 and PER2, 2 major components of the molecular clock, do not feature daily rhythmicity in bacterial composition and function.30 This indicates that information about the time of day is communicated to the microbiome by the host, and that this provides a means of synchronizing the activity of the meta-organism. It also indicates the co-evolutionary impact of the circadian clock, by demonstrating that diurnal rhythmicity can be achieved at several different levels of a prokaryotic-eukaryotic ecosystem (molecular and behavioral rhythmicity on the host side, community rhythmicity on the microbial side) and that coordination exists between these levels.
These findings also present a new perspective on the stability of the intestinal ecosystem. The described compositional and functional oscillations generate hour-scale fluctuations around a stable state that has so far been observed in longitudinal studies over longer periods of time. The dimension of day time should therefore be considered when interpreting studies of human and mouse microbiota composition. How the intestinal ecosystem orchestrates its rhythmic fluctuations and ensures a recurrent pattern in a 24-hour period remains elusive. In addition, the daily adaptations in oscillations around a stable composition might confer a large range of biological benefits to the ecosystem, including the prevention of pathogenic invasion of the microbial niche, and the detoxification of noxious xenobiotics. The introduction of such factors into the intestinal community is not constant over the course of a day, and anticipatory daily fluctuations in microbiome activity might be necessary to prevent detrimental effects on the ecosystem.
Dietary Determinants of Microbial Oscillations
A fundamental task of the circadian clock is the temporal orchestration of metabolism. Several interdependencies between the circadian clock and metabolic pathways have been identified, for instance the coupling of the NAD+ pathway to the core molecular clock.32-36 The cross-talk between metabolic pathways and the circadian clock ensures a temporal profiling of metabolic activities throughout the day. This tightly controlled sequence of metabolic events is necessary to synchronize the anabolic and catabolic pathways of energy turnover to diurnal variations in nutrient availability. Indeed, feeding time is a central driver of peripheral body clocks, as evidenced by 3 striking observations: First, timed feeding in which access to food is only provided at restricted times of the light-dark cycle uncouples peripheral clocks from the master clock in the brain, demonstrating autonomy of peripheral clocks and dominant entrainment by feeding times.37,38 Second, mice with genetic disruption of the circadian clock not only lose a large proportion of transcriptome oscillations, but also their strict nocturnal feeding pattern, indicating that the circadian clock governs the behavioral circuits determining rhythmic feeding-fasting activity. Third, timed feeding can, at least in part, restore this loss of transcriptional and behavioral rhythms, suggesting that oscillatory processes exist which solely rely on feeding rhythms, and can persist in the absence of a functional circadian clock.39,40
We found all 3 paradigms of food-entrainable clocks to apply to the intestinal microbiota. Limiting food access in mice to the dark or light phase only induced a phase shift in microbial oscillations while keeping the overall number of oscillatory commensals constant. Furthermore, the loss of bacterial oscillations in Per1/2-deficient mice, which are dysfunctional in the circadian clock, was restored by subjecting them to scheduled feeding.30 Therefore, feeding times are a dominant driver in the temporal orchestration of microbiome activity over the course of a day.
In addition to the timing of food intake, the type of food that is consumed seems to determine the activity of the circadian clock. Mice fed a high-fat diet exhibit attenuated amplitudes of clock gene oscillations, alterations in locomotor activity rhythms, and massive reprogramming of the circadian transcriptome.41,42 High-fat diet also alters the diurnal organization of the microbiome. While cyclic rhythmicity in the abundance of commensal bacteria generally persists in mice on high-fat diet, their overall number is reduced compared to controls, and cyclic bacteria species are not identical between the 2 conditions.31 The type of food is therefore a determinant of extent and magnitude of microbial community oscillations. Interestingly, time-restricted feeding prevents the adverse metabolic effects of high-fat feeding on the host,43,44 indicating that not the high-caloric, high-fat content of the diet per se, but rather the mistiming of nutrient availability with respect to circadian metabolic activity is responsible for the development of metabolic disease. Time-restricted feeding did not rescue the detrimental effect of high-fat feeding on diurnal oscillations in the intestinal microbiota; however, certain commensal species regained rhythmic activity upon scheduled feeding.31
Therefore, in addition to the entrainment of the host circadian system, feeding rhythmicity also exerts a powerful effect on the symbiotic community of bacteria colonizing the mammalian intestine (Fig. 1).
Linking Circadian Disruption, Metabolic Disease, and the Microbiome
Disruption of the circadian clock is a common hallmark of modern human lifestyle that was made possible with the invention of electric light, thereby creating independence of environmental light availability and uncoupling human activity from the diurnal timing of geophysical conditions. Together with the increase in global traveling activity across time zones, these changes have posed new challenges to the human circadian clock machinery, including repetitious re-adaptation to environmental light-dark conditions during jet lag and shift work. Chronic disruption of the circadian clock has been associated with a number of diseases, including obesity, diabetes, and cancer. Notably, these very conditions have been repeatedly associated with an aberrant composition of the intestinal microbiome, termed dysbiosis.45-49 Indeed, dysbiosis arises under conditions of clock deficiency, either in mice with genetic deletion of core clock components, or in humans and mice exposed to chronic environmentally induced circadian disturbances modeling jet lag or chronic shift work.30,50 This aberration of normal bacterial community composition seems to be functionally involved in the adverse metabolic consequences of clock disruption: Antibiotic treatment ameliorates obesity and glucose intolerance in mice subjected to jet lag, and transfer of dysbiosis from jet lagged mice and humans to germ-free mice fully transfers metabolic disease manifestations to a new host.30 These observations add circadian clock disruption to the list of conditions that can lead to microbe-induced obesity. They also suggest that the microbiota might constitute a previously unrecognized link between the rise of metabolic diseases and misalignment between the body clock and the geophysical time.
Perspective
The identification of diurnal microbiota oscillations provides a new perspective on our understanding of the interplay between the circadian clock and the environment. The rhythmic adaptation to geophysical fluctuations over the course of a day seems to be a functional principle that extends to the entire meta-organism of both host and symbiotic communities. Interestingly, both parts do not perform their rhythmic functions independently from one another, since the host circadian clock is required for diurnal oscillations of microbiota composition and function, and the presence of the microbiota is required for rhythmic signaling events in intestinal epithelial cells.51 The mechanisms of this novel aspect of host-microbiota interplay, the role of microbial oscillations for the beneficial effects of dietary interventions, and the connection of microbiota rhythmicity with the effects of the host circadian clock on metabolic health and longevity are intriguing aspects arising from the recognition of daily microbial cycles.
Furthermore, the diurnal program of the microbiome and its disruption in various settings of disease may offer an opportunity for nutritional and biotic interventions. Since feeding times are a major driver of microbial cycling, it might be possible to direct microbial activity to a preferred time of day. For instance, different diseases might be associated with exacerbated, diminished, or phase-shifted microbiome rhythms, and timed feeding might provide an elegant means of dietary intervention in order to restore healthy oscillations of commensals. Similarly, the impact of pro- and pre-biotic interventions on diurnal activity of the microbiome remains unknown, and will present an attractive avenue of future research on this new dimension of host-microbial interactions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the Elinav and Segal labs for fruitful discussions.
Funding
CAT is the recipient of a Boehringer Ingelheim Fonds PhD Fellowship. EE is supported by Yael and Rami Ungar, Israel; Abisch Frenkel Foundation for the Promotion of Life Sciences; the Gurwin Family Fund for Scientific Research; Leona M and Harry B Helmsley Charitable Trust; Crown Endowment Fund for Immunological Research; estate of Jack Gitlitz; estate of Lydia Hershkovich; the Benoziyo Endowment Fund for the Advancement of Science; Adelis Foundation; John L and Vera Schwartz, Pacific Palisades; Alan Markovitz, Canada; Cynthia Adelson, Canada; CNRS (Center National de la Recherche Scientifique); estate of Samuel and Alwyn J Weber; Mr. and Mrs. Donald L Schwarz, Sherman Oaks; grants funded by the European Research Council; the Kenneth Rainin Foundation; the German-Israel Binational foundation; the Israel Science Foundation; the Minerva Foundation; the Rising Tide foundation; and the Alon Foundation scholar award. EE is the incumbent of the Rina Gudinski Career Development Chair.
References
- 1. Bass J. Circadian topology of metabolism. Nature 2012. 491, 348-56; PMID: 23151577; http://dx.doi.org/ 10.1038/nature11704 [DOI] [PubMed] [Google Scholar]
- 2. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 2010. 72, 517-49; PMID:20148687; http://dx.doi.org/ 10.1146/annurev-physiol-021909-135821 [DOI] [PubMed] [Google Scholar]
- 3. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 2012. 35, 445-62; PMID:22483041; http://dx.doi.org/ 10.1146/annurev-neuro-060909-153128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & development 2000. 14, 2950-61; PMID:11114885; http://dx.doi.org/ 10.1101/gad.183500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science 2001. 291, 490-3; PMID:11161204; http://dx.doi.org/ 10.1126/science.291.5503.490 [DOI] [PubMed] [Google Scholar]
- 6. Panda S., et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002. 109, 307-20; PMID:12015981; http://dx.doi.org/ 10.1016/S0092-8674(02)00722-5 [DOI] [PubMed] [Google Scholar]
- 7. Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Ann Rev Biophys 2011. 40, 143-167; PMID:21332358; http://dx.doi.org/ 10.1146/annurev-iophys-042910-155317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature 2011. 469, 554-8,; PMID:21270895; http://dx.doi.org/ 10.1038/nature09654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teng SW, Mukherji S, Moffitt JR, de Buyl S, O'Shea EK. Robust circadian oscillations in growing cyanobacteria require transcriptional feedback. 2013. Science 340, 737-40,; PMID:23661759; http://dx.doi.org/ 10.1126/science.1230996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010. 464, 908-12; PMID:20376150; http://dx.doi.org/ 10.1038/nature08937 [DOI] [PubMed] [Google Scholar]
- 11. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. . Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013. 341, 1241214; PMID:24009397; http://dx.doi.org/ 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006. 444, 1027-31; PMID:17183312; http://dx.doi.org/ 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 13. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. . Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012. 143, 913-6 e917; PMID:22728514; http://dx.doi.org/ 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- 14. Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. . Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 2012. 150, 470-80; PMID:22863002; http://dx.doi.org/ 10.1016/j.cell.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A 2009. 106, 14728-33; PMID:19667173; http://dx.doi.org/ 10.1073/pnas.0904489106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suez J, Korem T, Zeevi D, Zilberman-Schapira G, Thaiss CA, Maza O, Israeli D, Zmora N, Gilad S, Weinberger A, et al. . Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 2014. 514, 181-6; PMID:25231862 [DOI] [PubMed] [Google Scholar]
- 17. Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, et al. . NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011. 145, 745-57; PMID:21565393; http://dx.doi.org/ 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rohlke F, Stollman N. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Therap Adv Gastroenterol 2012. 5, 403-20; PMID:23152734; http://dx.doi.org/ 10.1177/1756283X12453637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. . Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013. 155, 1451-63; PMID:24315484; http://dx.doi.org/ 10.1016/j.cell.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS biology 2007. 5, e177; http://dx.doi.org/ 10.1371/journal.pbio.0050177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwartz S, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, Dahl DB, Herman D, Wang M, Donovan SM, Chapkin RS. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. 2012. Genome biology 13, r32; PMID:22546241; http://dx.doi.org/ 10.1186/gb-2012-13-4-r32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scholtens PA, Oozeer R, Martin R, Amor KB, Knol J. The early settlers: intestinal microbiology in early life. Annu Rev Food Sci Technol 2012. 3, 425-47; PMID:22224552; http://dx.doi.org/ 10.1146/annurev-food-022811-101120 [DOI] [PubMed] [Google Scholar]
- 23. Sjogren YM, Jenmalm MC, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy 2009. 39, 518-26; PMID:19220322; http://dx.doi.org/ 10.1111/j.1365-2222.2008.03156.x [DOI] [PubMed] [Google Scholar]
- 24. Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. . The long-term stability of the human gut microbiota. Science 2013. 341, 1237439; PMID:23828941; http://dx.doi.org/ 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 2009. 9, 123; PMID:19508720; http://dx.doi.org/ 10.1186/1471-2180-9-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. . Human gut microbiome viewed across age and geography. 2012. Nature 486, 222-7; PMID:22699611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006. 444, 1022-3; PMID:17183309; http://dx.doi.org/ 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 28. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. . Linking long-term dietary patterns with gut microbial enterotypes. Science 2011. 334, 105-8; PMID:21885731; http://dx.doi.org/ 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014. 505, 559-63; PMID:24336217; http://dx.doi.org/ 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. . Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014. 159, 514-29; PMID:25417104; http://dx.doi.org/ 10.1016/j.cell.2014.09.048 [DOI] [PubMed] [Google Scholar]
- 31. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab 2014. 20, 1006-17; PMID:25470548; http://dx.doi.org/ 10.1016/j.cmet.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U, et al. . SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008. 134, 317-28; PMID:18662546; http://dx.doi.org/ 10.1016/j.cell.2008.06.050 [DOI] [PubMed] [Google Scholar]
- 33. Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008. 134, 329-40; PMID:18662547; http://dx.doi.org/ 10.1016/j.cell.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009. 324, 654-7; PMID:19286518; http://dx.doi.org/ 10.1126/science.1170803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. . Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009. 324, 651-4; PMID:19299583; http://dx.doi.org/ 10.1126/science.1171641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. . Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 2013. 342, 1243417; PMID:24051248; http://dx.doi.org/ 10.1126/science.1243417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mistlberger RE. Neurobiology of food anticipatory circadian rhythms. Physiology & behavior 2011. 104, 535-45; PMID:21527266; http://dx.doi.org/ 10.1016/j.physbeh.2011.04.015 [DOI] [PubMed] [Google Scholar]
- 38. Stephan FK, Swann JM, Sisk CL. Entrainment of circadian rhythms by feeding schedules in rats with suprachiasmatic lesions. Behav Neural Biol 25, 545-54 (1979; PMID:464989; http://dx.doi.org/ 10.1016/S0163-1047(79)90332-7 [DOI] [PubMed] [Google Scholar]
- 39. Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A 2009. 106, 21453-8; PMID:19940241; http://dx.doi.org/ 10.1073/pnas.0909591106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB, et al. . Harmonics of circadian gene transcription in mammals. PLoS Genet 2009. 5, e1000442; PMID:19343201; http://dx.doi.org/ 10.1371/journal.pgen.1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 2007. 6, 414-21; PMID:17983587; http://dx.doi.org/ 10.1016/j.cmet.2007.09.006 [DOI] [PubMed] [Google Scholar]
- 42. Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell 2013. 155, 1464-78; PMID:24360271; http://dx.doi.org/ 10.1016/j.cell.2013.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chaix A, Zarrinpar A, Miu P, Panda S. Time-restricted feeding is a preventative and therapeutic iIntervention against diverse nutritional challenges. Cell Metab 2014. 20, 991-1005; PMID:25470547; http://dx.doi.org/ 10.1016/j.cmet.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. . Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 2012. 15, 848-60; PMID:22608008; http://dx.doi.org/ 10.1016/j.cmet.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cox LM, Blaser MJ. Pathways in microbe-induced obesity. Cell Metab 2013. 17, 883-94; PMID:23747247; http://dx.doi.org/ 10.1016/j.cmet.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F, et al. . Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013. 498, 99-103; PMID:23719380; http://dx.doi.org/ 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
- 47. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, et al. . A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012. 490, 55-60; PMID:23023125; http://dx.doi.org/ 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 48. Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 2013. 13, 759-71; PMID:24154716; http://dx.doi.org/ 10.1038/nrc3611 [DOI] [PubMed] [Google Scholar]
- 49. Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014. 15, 317-28; PMID:24629338; http://dx.doi.org/ 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, Turek FW, Keshavarzian A. Circadian disorganization alters intestinal microbiota. PloS one 2014. 9, e97500; PMID:24848969; http://dx.doi.org/ 10.1371/journal.pone.0097500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 2013. 153, 812-27; PMID:23663780; http://dx.doi.org/ 10.1016/j.cell.2013.04.020 [DOI] [PubMed] [Google Scholar]


