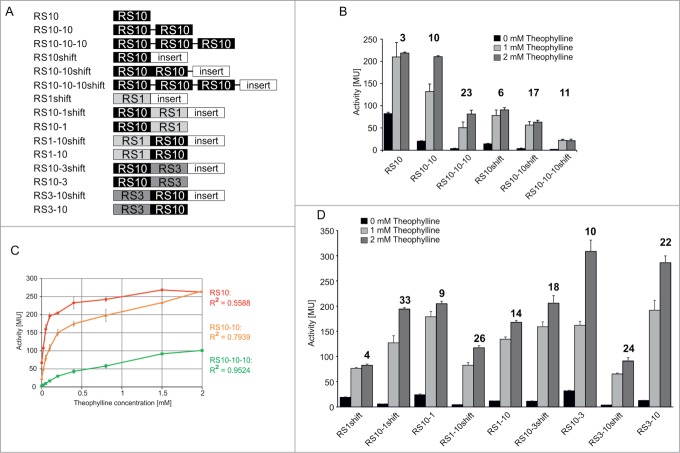
Figure 4.
Serial riboswitch constructs. (A) Schematic representation of tandem and tridem constructs upstream of the reporter gene (β-galactosidase or GFP), in 5′ to 3′ orientation. All transcripts start with a 3 nt-leader upstream of the first aptamer domain. For cloning reasons, several constructs carry a 6 nt-gap between the repeats, indicated by a black line. If no line is present, riboswitches directly follow each other. When indicated, an inert 19 nt region of the pBAD multiple cloning site (see ref.12) is inserted between riboswitch and Shine-Dalgarno sequence (RSshift constructs). The same arrangement was chosen for constructs with GFP reporter. (B) β-galactosidase activity of constructs with up to 3 serial riboswitch copies. While in the RS10 constructs the response ratio is increasing with additional riboswitch copies, the RS10–10–10shift construct shows a drop in activation compared to the corresponding tandem. (C) β-galactosidase activity as a function of theophylline concentration for the constructs RS10 (red), RS10–10 (orange) and RS10–10–10 (green). The linear regression coefficient R2 (colored accordingly) indicates that the linearity increases with the number of riboswitches. (D) β-galactosidase activity of tandem combinations of RS10 with RS1 or RS3, respectively. The approximate maximal activation ratio for all constructs is given as a number above each column set.