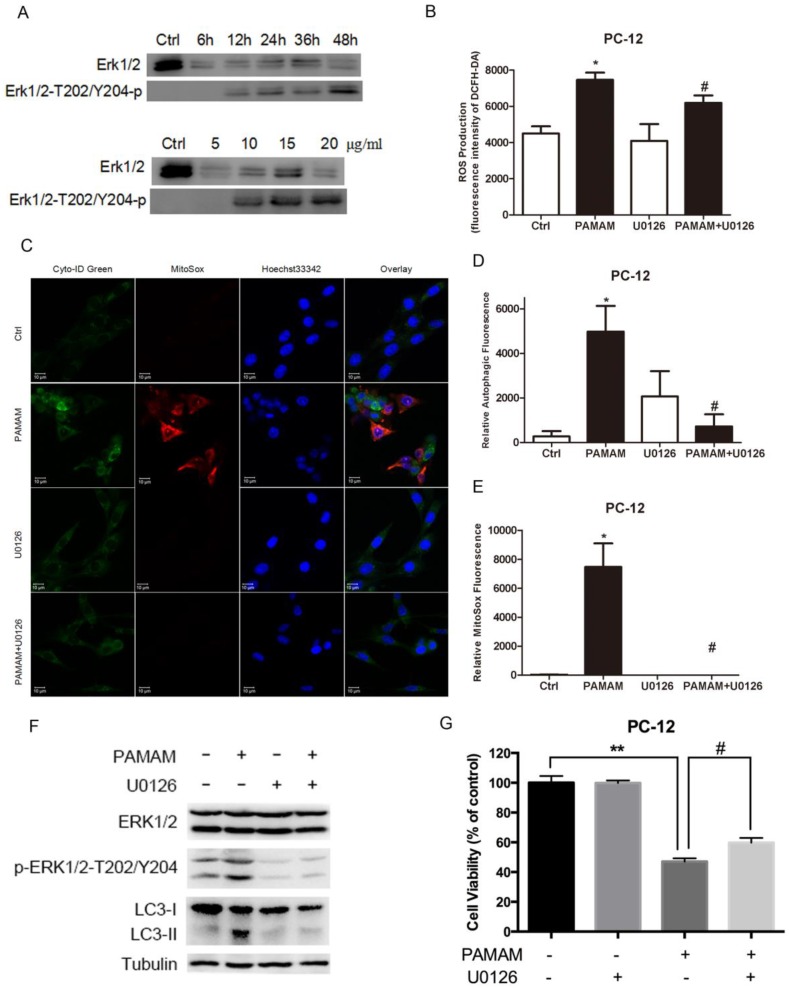
Figure 8.
ERK 1/2 signaling pathway was activated in oxidative stress and autophagy induced by PAMAM dendrimers. (A) PC-12 cells were seeded in a 6-well plate at an initial density of 2 × 105 cells per well. After 24 h of stabilization, cells were treated with 10 μg/mL of PAMAM dendrimers G5 for indicated times, or treated with different concentrations of PAMAM dendrimers G5 for 24 h. The same blot was used to probe Erk 1/2 and pErk 1/2. Changes in Erk 1/2 and pErk 1/2 were examined by Western blot. (B) PC-12 cells were incubated with or without PAMAM dendrimers in the presence or absence of U0126 for 24h. Cell samples were treated with DCFH-DA and analyzed by microplate reader to quantify levels of ROS. *p<0.05 versus Ctrl, and #p<0.05 versus PAMAM dendrimers G5-treated group. (C) PC-12 cells were treated as was described in (B). Cell samples were stained with Cyto-ID Green autophagy dye and MitoSox Red dye and analyzed by confocal microscopy. (D) and (E) The relative oxidative fluorescence intensity was quantified by confocal microscopy software. All data were presented as the means ± SD of more than three samples. *p<0.05 versus Ctrl, or versus PAMAM dendrimers G5-treated group. (F) Cells were treated as was described in (B), the whole protein was extracted, changes in Erk 1/2, pErk 1/2, and LC3 were analyzed by Western blot. (G) Cell growth inhibition was analyzed by CCK-8 assay. All data were presented as the means ± SD of four samples. *p<0.05 versus Ctrl, and #p<0.05 versus PAMAM dendrimers G5-treated group.