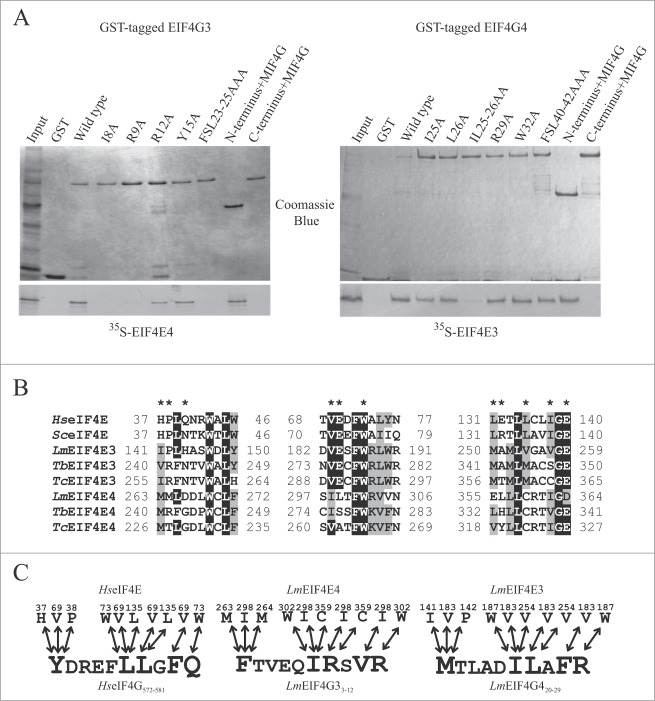
Figure 3.
Fine mapping of the interactions between L. major EIF4G3 and EIF4G4 with their eIF4E partners. (A) Fine mapping of the interactions between L. major EIF4G3 and EIF4E4 and between EIF4G4 and EIF4E3. Co-precipitation assays were carried out as described in Fig. 2 using either of the eIF4G homologues fusioned to GST and assayed for their ability to bind [35S]-labeled EIF4E3 and 4. The full-length eIF4G sequences as well as truncations lacking either the N or C-terminal regions of the proteins were used in the assay, as well as site-directed variants where selected residues were replaced by alanines as indicated. The results shown are representative of multiple co-precipitation assays carried out with a minimum of 2 independent sets of GST-tagged proteins. (B) Sequence alignment comparing the eIF4G binding residues (highlighted with a *), previously identified in 3 distinct blocs of the human and yeast eIF4E sequences,23,50 with the equivalent motifs found in different trypanosomatid EIF4E3 and 4 homologues. The alignment was carried out as described in Fig. 1 for the eIF4G homologues, but only the segments relevant for the eIF4G interaction are shown. (C) Schematic representation of either known or proposed interactions between different eIF4E/eIF4G homologues. The left scheme summarizes the interactions previously observed, based on the crystal structure,50 between human eIF4E and oligopeptides containing the consensus eIF4E binding motif from human eIF4G and eIF4E-binding proteins (YXXXXLΦ), plus the 3 subsequent residues. The middle and right schemes highlight the likely interactions presumed to occur between the trypanosomatid EIF4E4/EIF4G3 and EIF4E3/EIF4G4 pairs, assuming a conserved mode of binding between the different protein complexes.