Abstract
Interactions between protein and RNA play a key role in many biological processes in the gene expression pathway. Those interactions are mediated through a variety of RNA-binding protein domains, among them the highly abundant RNA recognition motif (RRM). Here we studied protein-RNA complexes from different RNA binding domain families solved by NMR and x-ray crystallography. Characterizing the structural properties of the RNA at the binding interfaces revealed an unexpected number of nucleotides with unusual RNA conformations, specifically found in RNA-RRM complexes. Moreover, we observed that the RNA nucleotides that are directly involved in interactions with the RRM domains, via hydrogen bonds and hydrophobic contacts, are significantly enriched with unique RNA conformations. Further examination of the sequences binding the RRM domain showed a preference for G nucleotides in syn conformation to precede or to follow U nucleotides in the anti-conformation, and U nucleotides in C2' endo conformation to precede U and G nucleotides possessing the more common C3' endo conformation. These findings imply a possible mode of RNA recognition by the RRM domains which enables the recognition of a wide variety of different RNA sequences and shapes. Overall, this study suggests an additional way by which the RRM domain recognizes its RNA target, involving a conformational readout.
Introduction
Protein-RNA interactions play a key role in all steps of the gene expression pathway. During co- and post transcription regulation RNA-binding proteins (RBPs) interact with the growing mRNA regulating its processing, transport and localization.1,2 More recently, it has been shown that RBPs also regulate many non-coding RNAs and are involved in their processing and escorting to their place of action.3 The interactions between RBPs and the RNA are mediated by a variety of protein domains. While single RNA-binding domains have been shown to be sufficient for RNA binding, many RBPs have a combination of multiple RNA-binding domains.4,5 RNA-binding domains can be classified into different subgroups according to their binding domain fold: αβ protein domains, zinc-finger and multimeric motifs. The most common αβ RNA-binding motifs are the RNA recognition motifs (RRMs),6-8 the K-homology (KH) domains9 that interact primarily with single-stranded RNA (ssRNA) and the double stranded RNA-binding domains (dsRBDs), which bind to double-stranded RNA (dsRNA).10 Among the αβ RNA-binding domains are also the Piwi Argonaut and Zwille (PAZ) domains that are found in proteins involved in the RNAi and microRNA processing pathway.11,12 Zinc-fingers (ZnF) are the most common nucleic-acids binding domains in eukaryotes. While the most common C2H2 ZnF domain is found mainly in DNA-binding proteins, the C2H2 and other ZnF domains have also been shown to bind RNA.13 Another common RNA binding domain is the Pumilio domain which is composed of multimeric repetitive motifs.14
During the last decade a large amount of structural data from X-ray crystallography and NMR, of protein-RNA complexes has been accumulating. Detailed analysis of the protein-RNA interactions of many of these complexes have demonstrated that RNA backbone interacts with the protein more frequently than the nucleotide bases, suggesting that the majority of RBP-RNA interactions are non-specific.15-20 Moreover, it has been shown that while in interactions via protruded surface the protein side chains often form electrostatic interaction with the backbone of the RNA in dented protein surfaces hydrogen bonds between the protein backbone and RNA bases are more frequent.21 The diverse structures of the RNA molecules may also serve as specific recognition sites allowing the bases to be more exposed for hydrogen bonding or for stacking interactions with the protein side chains.22-25 In addition, the RNA bases themselves can adopt different conformations.26,27 A possible role for the RNA conformation in RNA-protein recognition was suggested for the splicing factor SRSF2. It was shown that flipping the base conformation of 2 consecutive nucleotides allowed SRSF2 to bind 2 different RNA sequences equally well.28,29 A similar phenomenon was observed in the case of the HuD RNA binding protein.30 In previous studies, we demonstrated that ligand binding pockets on the ribosome are enriched in rare conformations of the nucleobase and the sugar pucker.31 The prevalence of syn conformation was also shown in active sites of functional RNA.32 Further, we demonstrated that nucleotides possessing the rare conformations are preferentially involved in direct interactions between RNA and small ligand.33 Here we show that rare RNA conformations are also prevalent among nucleotides which bind proteins, specifically via the RRM domain. This suggests that RNA conformation may contribute to the recognition code by which proteins can specifically identify their targets. Based on our results, we suggest that conformation readout has evolved only in specific protein families.
Materials and Methods
Data extraction
Structures of Protein-RNA complexes were obtained from the PDB, including structures that were solved either by X-ray crystallography (<=3.5Å) or NMR. Structures with polypeptide chains less than 40 amino acid residues and polyribonucleotide less than 5 nucleotides were removed. BLASTCLUST was employed to remove redundancy34 eliminating sequences which share more than 80% sequence identity. Further CLUSTALW35 was employed to align the nucleotides sequences within each cluster. Finally, a representative of the cluster with the longest RNA sequence was chosen. The PFAM database was used to classify domains.36 From each domain the protein-RNA interface residues were extracted using the Intervor web server37 excluding water molecules. Complexes including less than 5 nucleotides in the RNA interface were removed. For protein chains including more than one domain we excluded information from domains which had an overlap of >50 % of nucleotides in the binding interface, the domain with the smaller interface was removed. The final set included 198 domains (Supplementary File S1). The “All RNA” was extracted from Kligun et al.33
Structural properties calculations
Characterizing the structural properties of RNA within the interfaces was conducted by MC-annotate program.38 The features were extracted as described in39 using an in–house Perl script converting the MC-annotate output files into binary format, i.e., each nucleotide was given a score of “1” when a specific property was present and a score of “0” when it was absent. To calculate the relative abundance of a specific property, the fraction of nucleotides in the interface possessing the relevant property was calculated. The scores for each property were further standardized by the Z score, relative to either all RNA interfaces of the RNA-protein domains or to the background of all RNA (solved by the same technique X-ray or NMR). Clustering was performed using MeV software,40 applying K-Means clustering with Euclidean distance as the distance metric. The enrichment of specific domain families within the different clusters was evaluated using the chi square test for contingency table. Clusters which exhibited significant results were then analyzed using the Fisher's exact test to specify the statistical significance enrichment of each domain group in the cluster.
Analysis of RNA-protein interactions
Intermolecular hydrogen bonds and hydrophobic contacts were calculated using the HBPLUS program.41 The program computes all possible hydrogen atoms (H) between donor atoms (D) and acceptor atoms (A) that satisfy the distance and geometrical criteria for hydrogen bonding (see below). The distance criteria used to define a hydrogen bond were H-A distance <2.7Å, D-A distance <3.35Å and D-H-A angle >90°. Hydrophobic contacts were defined as all contacts between carbon atoms of the RNA and carbon atoms of the protein not involved in hydrogen bonds that were <3.9Å apart. The statistical significance of nucleotides in syn or C2' endo conformations to be involved in interactions between the RNA and the RRM domain relative to the general abundance in all other protein-RNA interactions was evaluated based on the hyper geometric distribution using the Fisher's exact test.
Results and Discussion
Rare RNA conformation in RNA-protein interfaces
To study the role of RNA conformation in RNA-protein recognition, we extracted a set of non-redundant structures of protein-RNA complexes from the PDB and classified the domains to different RNA binding domain families, as described in details in the Materials and Methods section. Further we extracted the RNA interfaces and compared the structural features of the RNA to the features extracted from a data set of all RNA interfaces of the RNA-binding domains. Consistent with the knowledge that most RBPs bind ssRNA33,42,43 we observed an overall preference for non-paired nucleotides in the RNA interfaces bound to the proteins, except for interfaces extracted from proteins belonging to the dsRBD family, which expectedly were enriched with the standard Watson-Crick pairing (WWcis). While for most RBPs we did not noticed any preference for a unique RNA-conformation at the RNA-protein interfaces, in some domain families, specifically in the RRM, we observed a significant enrichment for the rare RNA conformations, C2' endo and syn at the binding interface (Table 1). Further analysis of the nucleobases in the syn conformation showed a clear preference for purines (Table S1). These results are consistent with the fact that the rotation around the glycosidic bond from the common anti conformation to the less stable syn conformation, is more likely to occur in purines rather than in prymidines.44 This is also in agreement with a previous study that showed a preference for purines among syn nucleobases in functional RNAs.32 We also noticed that the C2' endo conformation, which was most significant in the RRM binding interfaces, was preferably found in U nucleotides (Table S1). It has been previously shown that the C2' endo unusual sugar pucker conformation provides less steric hindrance compared with the more common conformation C3' endo. Thus the C2' endo conformer is expected to be inherently more flexible, accommodating a wider range of allowed χ values and involving a lower energy cost specifically for bases in syn conformation.44-46 While the C2' endo conformation is relatively rare, it has previously been shown to play functionally important roles in RNA.47-50
Table 1.
Preferences for the different RNA conformations in RNA interfaces bound to different RNA-protein domains*
| C2' endo | C3' endo | Syn | anti | Non-paired | WW cis | |
|---|---|---|---|---|---|---|
| dsRBD | −0.66 | 0.77 | −0.48 | 0.54 | −1.42 | 1.52 |
| Helicase | −0.64 | 0.78 | −0.2 | −0.29 | 0.16 | −0.08 |
| ZnF | −0.4 | −0.59 | −0.52 | 0.58 | 0.03 | −0.04 |
| Piwi/Paz | −0.43 | 0.35 | −0.34 | 0.17 | 0.46 | −0.39 |
| Pumilio | −0.6 | 0.46 | −0.37 | 0.43 | 0.94 | −0.89 |
| S1/RNase | −0.29 | 0.32 | 0.01 | −0.05 | 0.73 | −0.7 |
| KH | 0.61 | −0.71 | 0.08 | −0.01 | 0.9 | −0.87 |
| RRM | 0.84 | −1.13 | 0.75 | −0.83 | 0.71 | −0.72 |
| Other | −0.06 | 0.28 | −0.09 | 0.15 | −0.4 | 0.36 |
For each family we calculated the fraction of nucleotides in the interface possessing the specific property (average of all the interfaces of the families) relative to the fraction in all RNA interfaces from all protein-RNA complexes. The scores for each property were further standardized by the Z score presented in the table.
Notably, since the complexes in our data set were solved by different techniques (X-ray crystallography and NMR) to ensure that the rare conformational properties are not biased by the technique used to solve the structure, we analyzed 100 randomly selected structures from the X-ray crystallography and from NMR. When calculating the frequency of the rare conformations in all structure we did not notice any significant differences between the 2 sets (P value < 0.38 for the syn conformation and P value < 0.2 for the C2' endo conformation, Mann-Whitney test). Overall, analysis of the subset of protein-RNA structures solved by X-ray crystallography with high resolution (better than 2.8Å) showed a very similar trend to what we found in the extended dataset (Table S2).
Unique structural properties of RRM-RNA interfaces
We further asked whether the RNA conformations and structural properties found in RNA interfaces associated with specific RNA-binding domains are characteristic properties of these domains. To this end, each interface was represented by a vector of properties normalized to a background of all RNA (Supplementary File S1). The interface vectors were then clusters using the K-means clustering (See Materials and Methods). As depicted in Figure 1A, the clustering resulted in 4 distinguished clusters. In Cluster 1 all interfaces were characterized by a preference for non-paired nucleotides with no enrichment of rare RNA conformations (Fig. 1A; Fig. S1). Analysis of the RNA binding proteins families comprising this cluster (Fig. 1B) showed no enrichment for a specific RNA-binding domain family. On the contrary, in cluster 2 (Fig. 1A; Fig. S1) which had an over representation of WW cis pairing, indeed, the primary family was the dsRBD (P value < 0.005, Fisher's exact test), which appears only in this cluster (Fig. 1B). The other group of interfaces in this cluster belonged to the ZnF domains that interact with dsRNA13,51 and to the helicase families, that are known to bind both ssRNA and dsRNA52,53 (Fig. 1B). The two remaining clusters (clusters 3 and 4) showed a significant enrichment for the rare RNA conformations (Fig. 1A; Fig. S1). As shown in Figure 1B the majority of the interfaces in this clusters belong to the RRM family (Fig. 1B), significantly enriched in cluster 3 (P value < 0.015, Fisher's exact test). Other domains represented in clusters 3 and 4 were mainly the S1/RNase and the KH (Fig. 1B). Interestingly, while RRM and S1 domain are not considered close homologs, both are comprised of 2-stranded β-sheet core which have been shown to contribute several conserved aromatic residues for stacking interactions with the nucleic acid bases.12,54,55 Nevertheless, the KH and the RRM share a similar fold and are considered the ancient RNA-binding domains.56,57 Furthermore, it has been shown that both domains recognize their RNA targets in a specific manner via amino acid side chains as well as via the main chain.43 In this study we found that the interfaces of both the RRM and the KH domains were clustered together, showing a significant preference for rare RNA conformations in their binding interfaces (Fig. 1A,B; Fig. S1).
Figure 1.
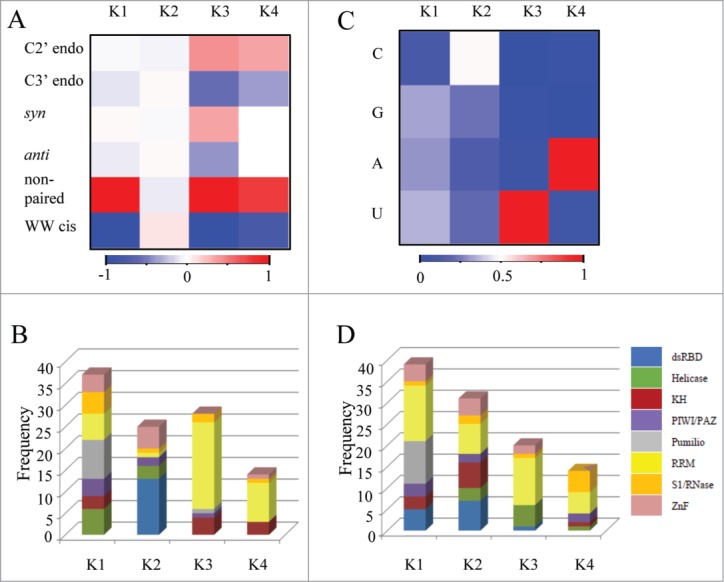
Clustering the RNAs from all protein-RNA complexes according to their different conformations and sequence features. The conformations and sequences of the RNAs extracted from all protein-RNA complexes were analyzed and represented as feature vectors. The vectors representing either the conformation or sequence signatures of the RNA from each complex were further clustered independently to 4 distinct clusters, employing the K-means clustering. (A) A heatmap representing the average frequency of each RNA conformation for all RNAs in each of the 4 clusters (B) The distribution of domain families within each of the 4 clusters that were classified according to the RNA conformations (C) A heatmap representing the average frequency of each of the 4 nucleobases in all RNA sequences within each of the 4 clusters (D) The distribution of domain families within each of the 4 clusters that were classified according to the nucleotide composition.
For comparison, we calculated the frequency of nucleotides in the interfaces and clustered the vectors using the K-means clustering (K = 4 ). As seen in Figure 1C, in clusters 1 and 2 the distribution of the 4 different nucleotides was relatively equal (Figs. 1C; Fig. S2), while cluster 3 was enriched with U (Fig. 1C; Fig. S2) and cluster 4 was enriched with A (Fig. 1C; Fig. S2). However, none of the clusters showed enrichment for a specific RNA binding domain family (Fig. 1D). These results reinforce that the enrichment of unique RNA conformation preferentially found in RRM and KH does not result from different nucleotides content of the RNA sequences bound to these protein families.
Enrichment of specific dinucleotides in RRM binding sequences
The observation that the RRM binding interfaces on RNA are enriched with rare RNA conformations led us to question whether the target sequences of proteins possessing the RRM domain are composed of unique compositions which tend to form the rare conformations. Given the limited amount of non-redundant data of protein-RNA complexes in the structural database we were not able to evaluate the statistical significance of all possible compositions, thus we concentrated on all possible dinucleotides possessing a rare conformation at either the first or the second nucleotide. Indeed, we found that the sequences that were extracted from RRM-RNA complexes were enriched for specific dinucleotides relative to the background of all RNA. Specifically, we observed an enrichment of G nucleotides in syn conformation to precede U nucleotides in anti-conformation (P value < 3.12e-08, Fisher's exact test) as well as G nucleotides in syn conformation to follow U nucleotides in anti-conformation (P value < 1.72e-05, Fisher's exact test) (Fig. 2A; Table S3A). In addition, we observed a preference for G nucleotides in syn conformation to precede A nucleotides in anti-conformation (P value < 0.0002, Fisher's exact test) and an unexpected enrichment of C nucleotides in syn conformation to precede U nucleotides in anti-conformation (P value < 0.0004, Fisher's exact test) (Fig. 2A; Table S3A). The three occurrences of CsynUanti were observed in 3 independent interactions between the RRM domains of the Polyprimidine Tract Binding protein (PTB) and the RNA (PDB codes 2AD9, 2ADB, 2ADC). Moreover we observed an enrichment of U nucleotides in C2' endo conformation to precede U nucleotides not in C2' endo conformation (P value < 3.18E-07, Fisher's exact test) and U nucleotides in C2' endo conformation to precede G nucleotides not in C2' endo conformation (P value < 3.77E-06, Fisher's exact test) (Fig. 2B; Table S3B). Further, we noticed a preference for U nucleotides in C2' endo conformation to follow G nucleotides not in C2' endo conformation (P value < 0.0009, Fisher's exact test) and A nucleotides in C2' endo conformation to follow A nucleotides not in C2' endo conformation (P value < 0.001, Fisher's exact test) (Fig. 2B; Table S3B). Taken together, we show that RNA sequences which are bound by the RRM domain are enriched for specific dinucleotides possessing unique RNA confor-mation. While as expected from the chemical nature of RNA nucleotides we noticed a preference for dinucleotides containing G in syn conformation44,32 and U in C2' endo conformation, we show that the RNA conformations were non-randomly distributed between all G and U containing dinucleotides, possibly providing an additional recognition code for RNA recognition by the RRM protein family.
Figure 2.
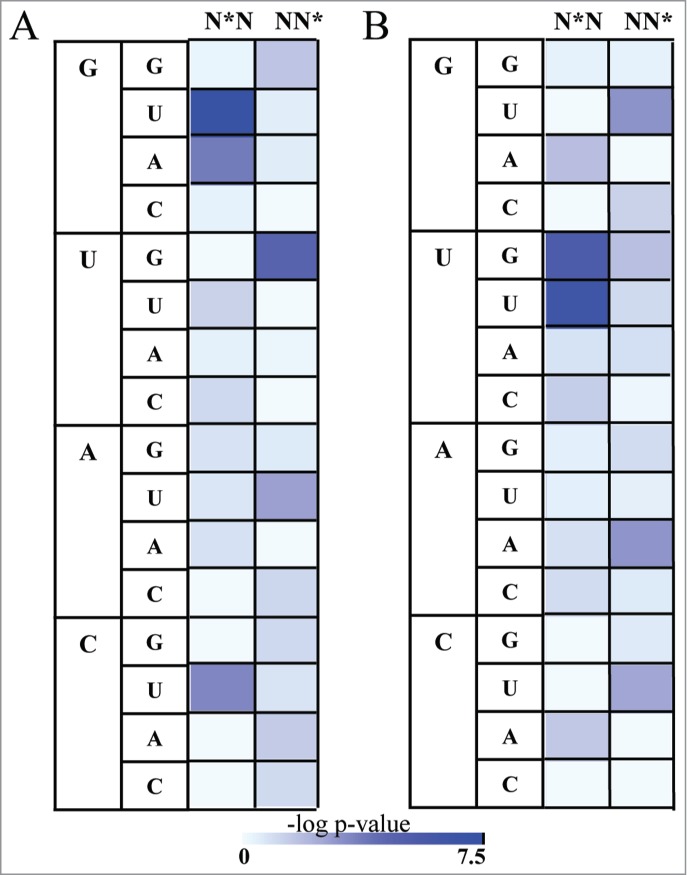
The distribution of Syn and C2' endo conformations in dinucleotides binding to the RRM domain. Asterix indicates nucleotides in rare conformation (syn or C2' endo) (A) Heatmap demonstrating the statistical significant of the enrichment of the dinucleotides possessing the syn conformations in RNA bound to RRM relative to the background of All RNA. The statistical significance is represented by the –log P value of the enrichment calculated using the Fisher's exact test following the Bonferroni correction, color range from azure (not significant) to blue (highly significant) (B) Heatmap demonstrating enrichment of the dinucleotides possessing the C2' endo conformations in RNA bound to RRM relative to the background of All RNA. The statistical significance is represented by the –log P value calculated using the Fisher's exact test following the Bonferroni correction, color range from azure (not significant) to blue (highly significant).
Unique RNA conformations are involved in direct interactions with the RRM
We further wanted to examine whether the enriched conformations are preferably found among the nucleotides directly interacting with the RRM amino acids. Zooming into the nucleotides involved in direct interactions with the RRM domain, we again found that the Syn conformation was significantly enriched in the RRM-RNA interactions compared to all protein-RNA interactions (Fig. 3). The latter was observed both when analyzing the interactions via hydrogen bonds (P value < 2.3e-09) and via hydrophobic contacts (P value < 2.2e-16). A very similar trend was observed for the rare C2' endo conformation (P value < 1.8e-05 for hydrogen bonds; P value < 2.2e-16 for hydrophobic contacts). Here again we found that interactions with the RRM domain preferably involve G nucleotides in syn conformations and U nucleotides in C2' endo conformation (Fig. 3; Tables S4, S5, S6, S7, and S8).
Figure 3.
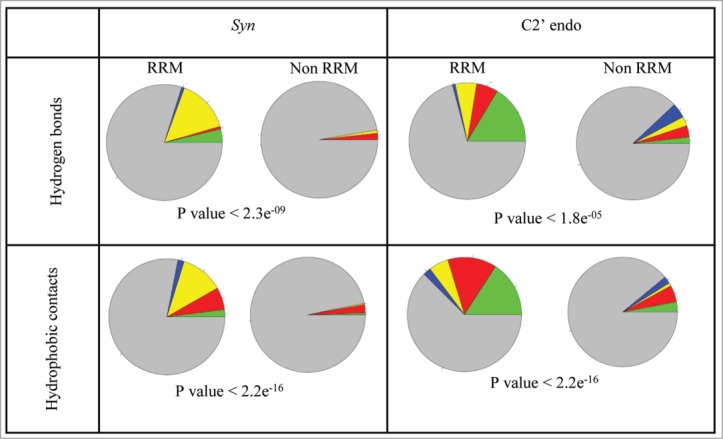
The preference of syn and C2' endo conformation in RRM-RNA interactions. Nucleotides in syn conformation or C2' endo conformation are presented as following: A are presented as red, U are presented as green, C are presented as blue and G are presented as yellow. Nucleotides in anti conformation or not in C2' endo conformation which are involved in the interactions are shown as gray. P value (Fisher's exact test) indicates the statistical preference of nucleotides in syn or C2' endo conformations to be involved in interactions between the RNA and the RRM domain, relative to the general abundance in all other protein-RNA interactions.
Further analysis of the interactions between nucleotides with rare RNA conformations and the amino acids revealed that G nucleotides in syn conformation is preferentially involved in hydrogen bond interactions with Arginine (Table S6C, Fig. 4B) and with Phenylalanine via hydrophobic contacts (Table S8C). In addition, we noticed a preference for U nucleotides in C2' endo conformation to contact Lysine via hydrogen bonds (Fig. 4C; Table S6B) and Phenylalanine via hydrophobic contacts (Table S8B). These results are consistent with the general preference of the positive amino acids to be involved in RNA-protein interactions.
Figure 4.
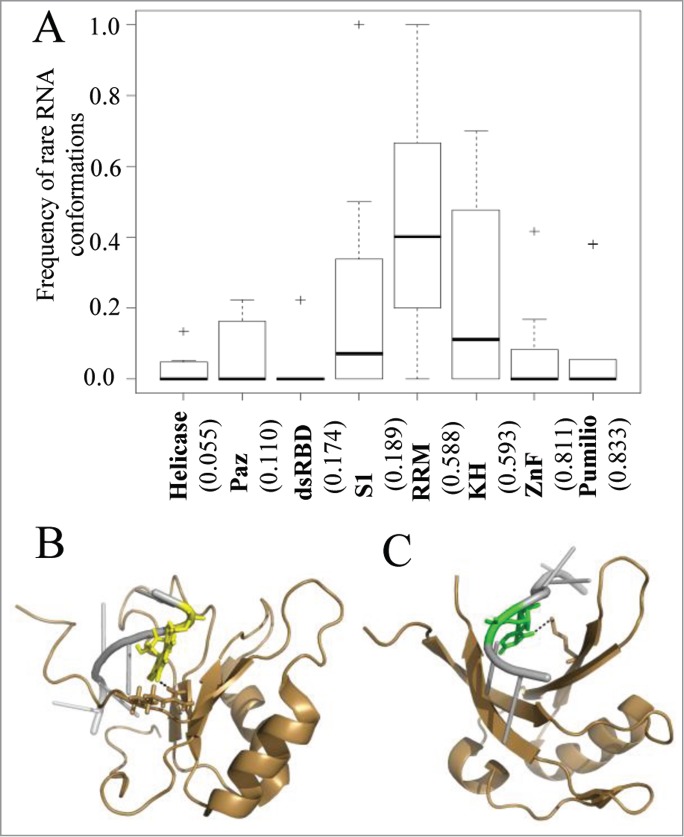
The interplay between sequence specificity and rare RNA conformations. (A) Box plots demonstrate the frequencies of interactions with rare RNA conformations in RNAs from protein-RNA complexes grouped according the interacting RNA binding domain family. The numbers in the x-axis below each plot indicate the calculated average frequency of specific hydrogen bond interactions within each family. In (B and C) are examples of RRM-RNA complexes from the PDB which involve hydrogen bond interactions between the RRM domain and nucleotides with rare conformations. The RNA and protein are colored in silver and sand, respectively. Hydrogen bonds between the protein domain and the RNA are depicted in dashed black lines. (B) Graphical representation of the interactions of the RRM domain with nucleotide in syn conformation (PDB ID 2LEC). Highlighted are the interacting pair G (yellow) and ARG (stick). (C) Graphical representation of the interactions of the RRM domain with nucleotide in C2' endo conformation (PDB ID 2XS2). Highlighted are the interacting pair U (green) and LYS (stick).
We further wanted to explore whether amino acids in specific positions in the RNA-binding domains are preferentially involved in interactions with nucleotides in rare RNA conformations. When analyzing the interactions associated with the RRM domain we did not observe any preference for an amino acid in a specific location in the binding domain to be involved in interactions with a nucleotide possessing a rare conformation. Overall, most of the interactions with nucleotides in rare conformations involved amino acids located in β sheets, consistent with the well-established knowledge that the primary RNA binding surface of the RRM is the 4-stranded β-sheet.43 While the amino acids involved in interactions with the rare nucleotides in RNA via hydrogen bonds were found in β2 and β4 of the RRM domain, the amino acids involved in hydrophobic contacts were mostly found in β1 and β3 (Tables S9A and S10A). However, as shown in Supplemental Tables 9A and 10A, there was no significant difference between the distribution of amino acids involved in interactions with all RNA nucleotides to the distribution of the amino acids involved in interactions with nucleotides in rare conformations only. In addition to the conserved αβ fold of the RRM, this domain is also characterized by 2 conserved sequences, RNP1 and RNP2.8,43 We further tested whether the amino acids interacting with rare conformations are preferentially located in one of these conserved regions, but again we did not observe any significant preferences (Tables S9B and S10B). Moreover, while we did observe that the 3 conserved aromatic side-chains in the 2 central β strands (β1 and β3) of the RRM, known to be involved in interactions with positions N1 and N2 in the RNA target sequence,8,43 are commonly involved in aromatic interactions with nucleotides in rare conformations, there was no indication for preferential binding of one of these conserved residues to nucleotides in rare conformations (Table S10C). Consistently, when exploring the interactions of amino acids in the KH domain to nucleotides with unique RNA conformational, we did not notice any significant preferences for residues located at specific regions of the KH domain9,43 to be involved in direct interactions with nucleotides in rare conformations (Table S11).
RNA sequence-specific recognition versus RNA conformational recognition in RNA-proteins families
Sequence-specific recognition modes were characterized for many RBP families, including the pumilio,14,58 ZnF,59 KH9 and the RRM.60 In most of these families sequence specificity is achieved primarily via direct interactions between the amino acids side chains and the RNA base edges keeping the sugar-phosphate backbone exposed to the solvent.43 This type of binding differs from the binding of non- sequence specific RNA binding proteins to ssRNA (e.g helicases)61 that is mainly mediated by positively charged side-chains that contact the sugar-phosphate backbone of the RNA, while the RNA bases are exposed to the solvent.43 The binding of dsRBD has also been described as non-sequence specific, binding preferentially to a double stranded A-form RNA helix. Recent structural studies of dsRBPs have also demonstrated a direct readout of RNA sequence in the minor groove of the helix.10
To investigate the relationship between RNA conformation and sequence-specific recognition in protein-RNA recognition, we analyzed the frequency of interactions with rare RNA conformations relative to the frequency of specific interactions (defined as the percent of hydrogen bonds between the amino acid side chains and the RNA base atoms from the total of all hydrogen bonds interactions). As can be noticed from Figure 4A, there is no correlation between the fraction of interactions involving nucleotides with rare conformational and the normalized frequency of sequence-specific interactions. While Znf and Pumilio domains which are characterized by high sequence specific interactions have very few interactions with nucleotides with unusual conformations, other domains with similar levels of sequence specific interactions, such as the RRM and KH were highly enriched with interactions via rare RNA conformations. On the contrary, both domains that showed low level of interactions via rare RNA conformations (the Helicase, Paz and dsRBD) and domain which are characterized by interactions via rare RNA conformations (such as S1) do not preferably bind their RNA targets via specific interactions.
Overall, our results suggest that RNA conformational provides an additional level by which RNA sequences can be recognized by RBPs. This recognition mode is mainly attributed to the RRM protein family, but also found in other families like the KH and the S1 domains with no correlation to the otherwise preference of these domains to bind the RNA via specific or non-specific interactions.
Conclusions
In this study we explore the possible role of unique RNA conformation (specifically syn conformation and C2'endo conformations) in specific RNA recognition by proteins. Our comprehensive analysis of the RNA conformations in a broad non-redundant set of RNA-proteins 3D complexes solved by either X-ray crystallography or NMR revealed that while rare RNA conformation is not highly frequent in all RNA-protein complexes, it is significantly enriched in sequences bound to the RRM. Further, examination of the sequences binding the RRM domain showed a preference for specific dinucleotides containing the rare conformations. The overall enrichment of rare RNA conformations in RRM-RNA complexes was even more profound when looking at the subset of nucleotides directly involved in interactions with the protein. Overall, our results shed new light on RNA recognition in the most widespread RNA-binding domain in eukaryotes, suggesting a new mode of recognition, possibly explaining the highly diverse range of sequences which are bound via this domain.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Stefl R, Skrisovska L, Allain FH. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep 2005; 6:33-8; PMID:15643449; http://dx.doi.org/ 10.1038/sj.embor.7400325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008; 582:1977-86; PMID:18342629; http://dx.doi.org/ 10.1016/j.febslet.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masuda K, Kuwano Y, Nishida K, Rokutan K, Imoto I. NF90 in posttranscriptional gene regulation and microRNA biogenesis. Int J Mol Sci 2013; 14:17111-21; PMID:23965975; http://dx.doi.org/ 10.3390/ijms140817111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ankö ML, Neugebauer KM. RNA-protein interactions in vivo: global gets specific. Trends Biochem Sci 2012; 37:255-62; PMID:22425269; http://dx.doi.org/ 10.1016/j.tibs.2012.02.005 [DOI] [PubMed] [Google Scholar]
- 5.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet 2014; 15:829-45; PMID:25365966; http://dx.doi.org/ 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 2008; 18:290-8; http://dx.doi.org/ 10.1016/j.sbi.2008.04.002 [DOI] [PubMed] [Google Scholar]
- 7.Muto Y, Yokoyama S. Structural insight into RNA recognition motifs: versatile molecular Lego building blocks for biological systems. Wiley Interdiscip Rev RNA 2012; 3:229-46; PMID:22278943; http://dx.doi.org/ 10.1002/wrna.1107 [DOI] [PubMed] [Google Scholar]
- 8.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 2005; 272:2118-31; PMID:15853797; http://dx.doi.org/ 10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- 9.Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J 2008; 275:2712-26; PMID:18422648; http://dx.doi.org/ 10.1111/j.1742-4658.2008.06411.x [DOI] [PubMed] [Google Scholar]
- 10.Masliah G, Barraud P, Allain FH. RNA recognition by double-stranded RNA binding domains: a matter of shape and sequence. Cell Mol Life Sci 2013; 70:1875-95; PMID:22918483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Varani G. Protein families and RNA recognition. FEBS J 2005; 272:2088-97; PMID:15853794; http://dx.doi.org/ 10.1111/j.1742-4658.2005.04650.x [DOI] [PubMed] [Google Scholar]
- 12.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol 2007; 8:479-90; PMID:17473849; http://dx.doi.org/ 10.1038/nrm2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown RS. Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol 2005; 15:94-8; PMID:15718139; http://dx.doi.org/ 10.1016/j.sbi.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 14.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell 2002; 110:501-12; PMID:12202039; http://dx.doi.org/ 10.1016/S0092-8674(02)00873-5 [DOI] [PubMed] [Google Scholar]
- 15.Treger M, Westhof E. Statistical analysis of atomic contacts at RNA-protein interfaces. J Mol Recognit 2001; 14:199-214; PMID:11500966; http://dx.doi.org/ 10.1002/jmr.534 [DOI] [PubMed] [Google Scholar]
- 16.Jeong E, Kim H, Lee SW, Han K. Discovering the interaction propensities of amino acids and nucleotides from protein-RNA complexes. Mol Cells 2003; 16:161-7; PMID:14651256 [PubMed] [Google Scholar]
- 17.Phipps KR, Li H. Protein-RNA contacts at crystal packing surfaces. Proteins 2007; 67:121-7; PMID:17211891; http://dx.doi.org/ 10.1002/prot.21230 [DOI] [PubMed] [Google Scholar]
- 18.Ellis JJ, Broom M, Jones S. Protein-RNA interactions: structural analysis and functional classes. Proteins 2007; 66:903-11; PMID:17186525; http://dx.doi.org/ 10.1002/prot.21211 [DOI] [PubMed] [Google Scholar]
- 19.Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res 2008; 36:2705-16; PMID:18353859; http://dx.doi.org/ 10.1093/nar/gkn102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Gribskov M. The role of RNA sequence and structure in RNA–protein interactions. J Mol Biol 2011; 409:574-87; PMID:21514302; http://dx.doi.org/ 10.1016/j.jmb.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 21.Iwakiri J, Tateishi H, Chakraborty A, Patil P, Kenmochi N. Dissecting the protein-RNA interface: the role of protein surface shapes and RNA secondary structures in protein-RNA recognition. Nucleic Acids Res 2012; 40:3299-306; PMID:22199255; http://dx.doi.org/ 10.1093/nar/gkr1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagai K. RNA-protein complexes. Curr Opin Struct Biol 1996; 6:53-61; PMID:8696973; http://dx.doi.org/ 10.1016/S0959-440X(96)80095-9 [DOI] [PubMed] [Google Scholar]
- 23.Jones S, Daley DT, Luscombe NM, Berman HM, Thornton JM. Protein-RNA interactions: a structural analysis. Nucleic Acids Res 2001; 29:943-54; PMID:11160927; http://dx.doi.org/ 10.1093/nar/29.4.943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cusack S. RNA-protein complexes. Curr Opin Struct Biol 1999; 9:66-73; PMID:10400475; http://dx.doi.org/ 10.1016/S0959-440X(99)80009-8 [DOI] [PubMed] [Google Scholar]
- 25.Valegård K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature 1994; 371:623-6; http://dx.doi.org/ 10.1038/371623a0 [DOI] [PubMed] [Google Scholar]
- 26.Murthy VL, Srinivasan R, Draper DE, Rose GD. A complete conformational map for RNA. J Mol Biol 1999; 291:313-27; PMID:10438623; http://dx.doi.org/ 10.1006/jmbi.1999.2958 [DOI] [PubMed] [Google Scholar]
- 27.Schneider B, Morávek Z, Berman HM. RNA conformational classes. Nucleic Acids Res 2004; 32:1666-77; PMID:15016910; http://dx.doi.org/ 10.1093/nar/gkh333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daubner GM, Cléry A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J 2012; 31:162-74; PMID:22002536; http://dx.doi.org/ 10.1038/emboj.2011.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daubner GM, Cléry A, Allain FH. RRM-RNA recognition: NMR or crystallography…and new findings. Curr Opin Struct Biol 2013; 23:100-8; PMID:23253355; http://dx.doi.org/ 10.1016/j.sbi.2012.11.006 [DOI] [PubMed] [Google Scholar]
- 30.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat Struct Biol 2001; 8:141-5; PMID:11175903; http://dx.doi.org/ 10.1038/84131 [DOI] [PubMed] [Google Scholar]
- 31.David-Eden H, Mankin AS, Mandel-Gutfreund Y. Structural signatures of antibiotic binding sites on the ribosome. Nucleic Acids Res 2010; 38:5982-94; PMID:20494981; http://dx.doi.org/ 10.1093/nar/gkq411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sokoloski JE, Godfrey SA, Dombrowski SE, Bevilacqua PC. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA 2011; 17:1775-87; PMID:21873463; http://dx.doi.org/ 10.1261/rna.2759911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kligun E, Mandel-Gutfreund Y. Conformational readout of RNA by small ligands. RNA Biol 2013; 10:982-90; PMID:23618839; http://dx.doi.org/ 10.4161/rna.24682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10; PMID:2231712; http://dx.doi.org/ 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673-80; PMID:7984417; http://dx.doi.org/ 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al.. The Pfam protein families database. Nucleic Acids Res 2012; 40:D290-301; PMID:22127870; http://dx.doi.org/ 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cazals F, Proust F, Bahadur RP, Janin J. Revisiting the Voronoi description of protein-protein interfaces. Protein Sci 2006; 15:2082-92; PMID:16943442; http://dx.doi.org/ 10.1110/ps.062245906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gendron P, Lemieux S, Major F. Quantitative analysis of nucleic acid three-dimensional structures. J Mol Biol 2001; 308:919-36; PMID:11352582; http://dx.doi.org/ 10.1006/jmbi.2001.4626 [DOI] [PubMed] [Google Scholar]
- 39.Banatao DR, Altman RB, Klein TE. Microenvironment analysis and identification of magnesium binding sites in RNA. Nucleic Acids Res 2003; 31:4450-60; PMID:12888505; http://dx.doi.org/ 10.1093/nar/gkg471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol 2006; 411:134-93; PMID:16939790; http://dx.doi.org/ 10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
- 41.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol 1994; 238:777-93; PMID:8182748; http://dx.doi.org/ 10.1006/jmbi.1994.1334 [DOI] [PubMed] [Google Scholar]
- 42.Messias AC, Sattler M. Structural basis of single-stranded RNA recognition. Acc Chem Res 2004; 37:279-87; PMID:15147168; http://dx.doi.org/ 10.1021/ar030034m [DOI] [PubMed] [Google Scholar]
- 43.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res 2006; 34:4943-59; PMID:16982642; http://dx.doi.org/ 10.1093/nar/gkl620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bloomfield VA, Crothers DM, Ignacio Tinoco J. Nucleic Acids: Structures, properties, and functions. University Science Books, 2000. [Google Scholar]
- 45.Kowalak JA, Bruenger E, McCloskey JA. Posttranscriptional modification of the central loop of domain V in Escherichia coli 23 S ribosomal RNA. J Biol Chem 1995; 270:17758-64; PMID:7629075; http://dx.doi.org/ 10.1074/jbc.270.30.17758 [DOI] [PubMed] [Google Scholar]
- 46.Dalluge JJ, Hashizume T, Sopchik AE, McCloskey JA, Davis DR. Conformational flexibility in RNA: the role of dihydrouridine. Nucleic Acids Res 1996; 24:1073-9; PMID:8604341; http://dx.doi.org/ 10.1093/nar/24.6.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mortimer SA, Weeks KM. C2'-endo nucleotides as molecular timers suggested by the folding of an RNA domain. Proc Natl Acad Sci U S A 2009; 106:15622-7; PMID:19717440; http://dx.doi.org/ 10.1073/pnas.0901319106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaGrandeur TE, Hüttenhofer A, Noller HF, Pace NR. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J 1994; 13:3945-52; PMID:7521296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leontis NB, Westhof E. A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs. J Mol Biol 1998; 283:571-83; PMID:9784367; http://dx.doi.org/ 10.1006/jmbi.1998.2106 [DOI] [PubMed] [Google Scholar]
- 50.Kolev NG, Steitz JA. In vivo assembly of functional U7 snRNP requires RNA backbone flexibility within the Sm-binding site. Nat Struct Mol Biol 2006; 13:347-53; PMID:16547514; http://dx.doi.org/ 10.1038/nsmb1075 [DOI] [PubMed] [Google Scholar]
- 51.Hall TM. Multiple modes of RNA recognition by zinc finger proteins. Curr Opin Struct Biol 2005; 15:367-73; PMID:15963892; http://dx.doi.org/ 10.1016/j.sbi.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 52.Lüking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev Biochem Mol Biol 1998; 33:259-96; PMID:9747670; http://dx.doi.org/ 10.1080/10409239891204233 [DOI] [PubMed] [Google Scholar]
- 53.Marintchev A. Roles of helicases in translation initiation: a mechanistic view. Biochim Biophys Acta 2013; 1829:799-809; PMID:23337854; http://dx.doi.org/ 10.1016/j.bbagrm.2013.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell 1997; 88:235-42; PMID:9008164; http://dx.doi.org/ 10.1016/S0092-8674(00)81844-9 [DOI] [PubMed] [Google Scholar]
- 55.Schubert M, Edge RE, Lario P, Cook MA, Strynadka NC, Mackie GA, McIntosh LP. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J Mol Biol 2004; 341:37-54; PMID:15312761; http://dx.doi.org/ 10.1016/j.jmb.2004.05.061 [DOI] [PubMed] [Google Scholar]
- 56.Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science 1994; 265:615-21; PMID:8036511; http://dx.doi.org/ 10.1126/science.8036511 [DOI] [PubMed] [Google Scholar]
- 57.Lorković ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 2002; 30:623-35; PMID:11809873; http://dx.doi.org/ 10.1093/nar/30.3.623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Varani G. Finding the missing code of RNA recognition by PUF proteins. Chem Biol 2011; 18:821-3; PMID:21802002; http://dx.doi.org/ 10.1016/j.chembiol.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 2004; 11:257-64; PMID:14981510; http://dx.doi.org/ 10.1038/nsmb738 [DOI] [PubMed] [Google Scholar]
- 60.Allers J, Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J Mol Biol 2001; 311:75-86; PMID:11469858; http://dx.doi.org/ 10.1006/jmbi.2001.4857 [DOI] [PubMed] [Google Scholar]
- 61.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 2006; 125:287-300; PMID:16630817; http://dx.doi.org/ 10.1016/j.cell.2006.01.054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


