Figure 2.
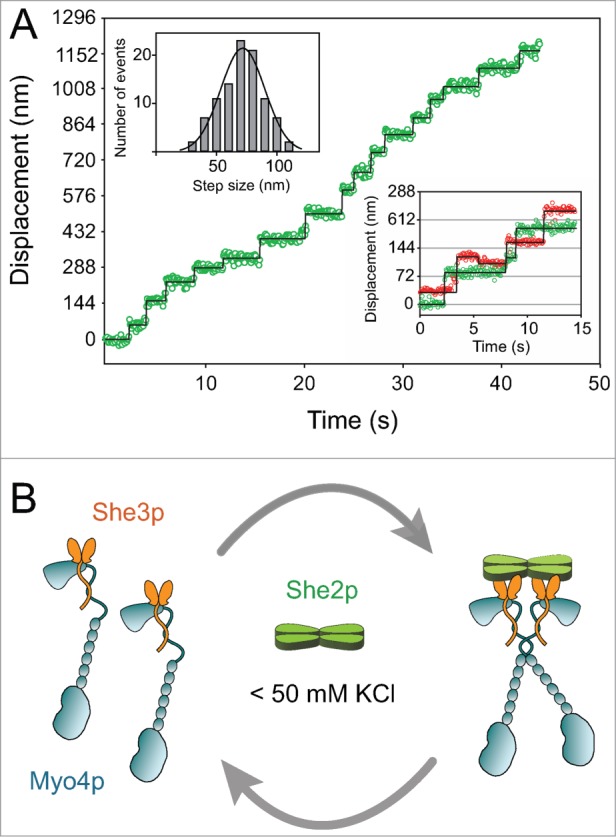
(A) Trace showing the displacement vs. time of the Myo4p–She3p–She2p motor complex on an actin filament at low ionic strength (50 mM KCl). The multiple steps without dissociation illustrate that the complex is processive. Myo4p–She3p is labeled with a Quantum dot (Qdot) on only one head of the two-headed motor complex. Using this labeling strategy, the head position of myosin can be tracked on actin with high time and spatial resolution by fitting the Qdot image of each frame to a two-dimensional Gaussian, which approximates its point spread function and allows its x and y positions to be determined with ∼6 nm resolution. The center of a distribution can be determined much more accurately than the width, which is limited by the ∼200 nm resolution of a light microscope. The top left inset shows the histogram of step sizes, that average 72 ± 18 nm. The bottom right inset shows a stepping trace of a two-headed complex in which each head is labeled with a different colored Quantum dot. These data illustrate the hand-over-hand stepping pattern of the two heads, which implies communication between the heads. (B) Diagram showing that She2p recruits two single-headed Myo4p–She3p complexes to form a processive double-headed motor complex at low ionic strength (< 50 mM KCl). She3p is shown as a dimer interacting with the Myo4p globular tail and rod.
