Figure 7.
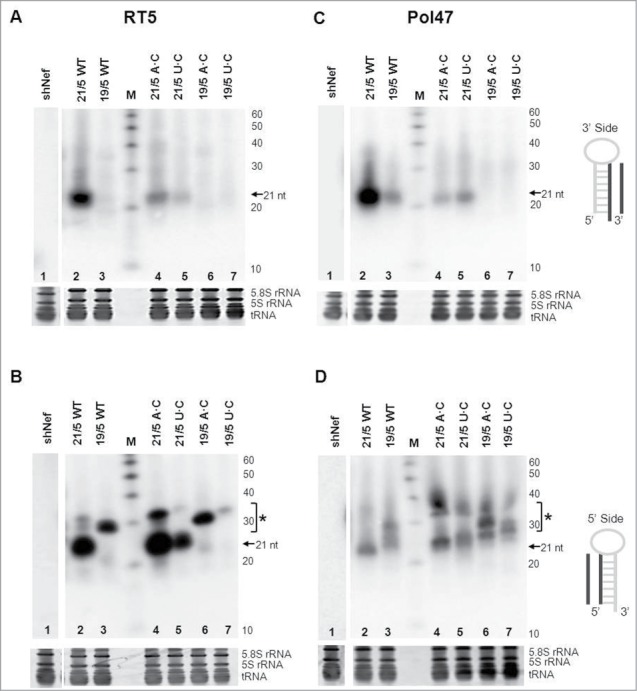
Processing of shRNAs and AgoshRNAs is influenced by the bottom base pair. (A, B) RT5 and (C, D) Pol47 variants were analyzed by RNA gel blot. HEK293T cells were transfected with 5 μg of the corresponding RNA constructs. The shRNAs and AgoshRNAs (21/5 and 19/5) varied in bottom bp (WT is A-U for RT5 and G-C for Pol47, and mutants A·C or U·C). Total RNA was isolated and analyzed by northern blot using an LNA probe to detect processing products derived from the 3′ side (A, C) and the 5′ side (B, D). Size markers were included (length indicated in nt). An unrelated shRNA (shNef) was included as negative control. The regular shRNA ∼21 nt products are marked and * indicates the AgoshRNA ∼30 nt products.
