Fig. 2.
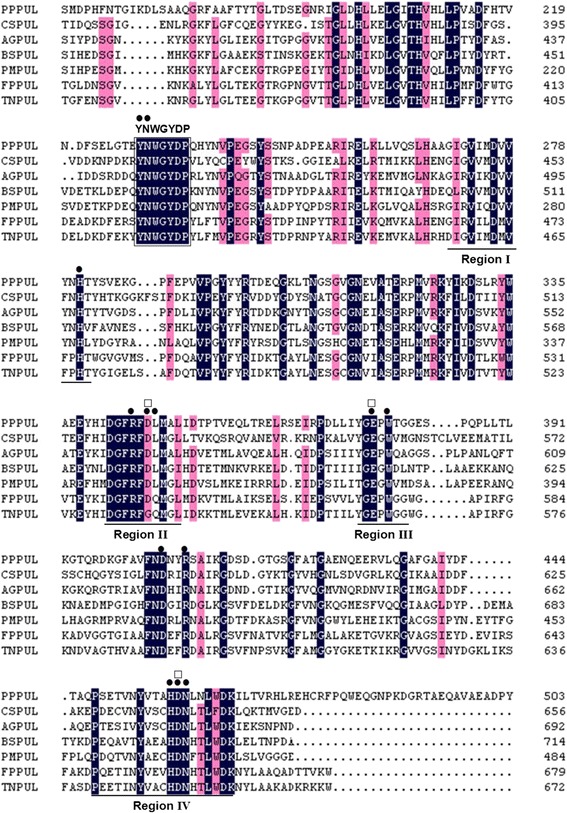
Sequence alignment of the conserved region of PulN and pullulanase Type I from other different bacterial sources. The sequences used in this alignment were obtained from GenBank as following. PPPUL: P. polymyxa pullulanase (this study; GenBank accession no. KJ740392); CSPUL: C. saccharolyticus pullulanase (GenBank accession no. L39876); AGPUL: A. gottschalkii pullulanase (GenBank accession no. AY541591); BSPUL: Bacillus sp. CICIM 263 pullulanase (GenBank accession no. AGA03915); PMPUL: P. mucilaginosus KNP414 pullulanase (GenBank accession no. YP004645603); FPPUL: F. pennavorans Ven5 pullulanase (GenBank accession no. AF096862); TNFUL: T. neapolitana strain KCCM 41025 pullulanase (GenBank accession no. FJ716701). Regions I, II, III, and IV are lined. YNWGYDP conserved in all type I pullulanases is boxed. The numbering refers to the amino acid position in each sequence. The structures are denoted as follows: ●, the active site; □, the catalytic site
