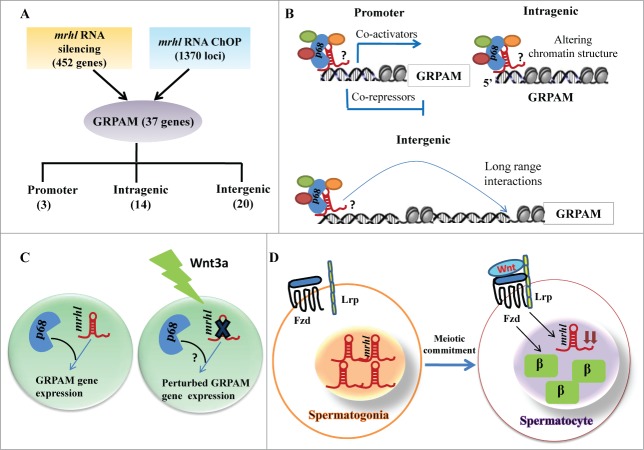
Figure 12.
Summary of the relationship between chromatin occupancy of mrhl RNA and regulation of gene expression in the context of Wnt signaling in spermatogenesis. (A) Overlap set of mrhl RNA silencing (microarray data) and mrhl RNA ChOP leading to 37 GRPAM genes and their classification based on location of ChOP sequence reads. Number in brackets indicates the number of GRPAM of that particular category. (B) Hypothetical mechanisms of gene regulation by mrhl RNA-p68 complex at GRPAM loci. Oval circles indicate different proteins associated with mrhl RNA which can include co-activators or co-repressors or chromatin modifiers. ? indicates that the nature of interaction of p68-mrhl RNA complex with chromatin template is not clear at present. Regulation of promoter class of GRPAM can be through recruitment of co-activators or co-repressors while intragenic and intergenic GRPAM can be regulated through alteration of chromatin structure and long range interactions respectively. (C) Down regulation of mrhl RNA and perturbation of GRPAM gene expression in Gc1-Spg spermatogonial cell line upon Wnt3a treatment. ? indicates that the status of p68 genome wide occupancy and expression of p68 upon Wnt3a treatment is not known. (D) Inverse correlation between Wnt activated state (as demonstrated by nuclear β catenin) and mrhl RNA expression levels between spermatogonia (Repressed Wnt signaling, higher expression of mrhl RNA) and differentiated spermatocytes (Wnt activation and decreased levels of mrhl RNA ). β: β catenin, Fzd: Frizzled, Lrp: Low-density lipoprotein receptor-related protein.