Figure 1.
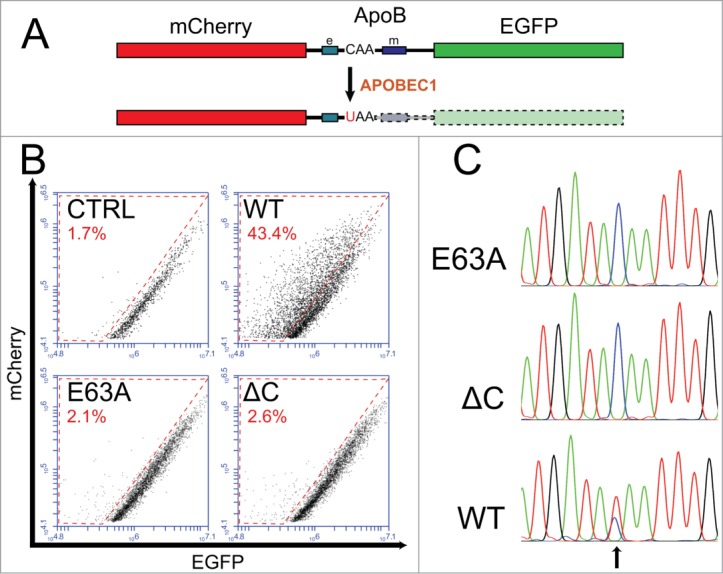
APOBEC1 edits the mRNA of a chimeric construct containing the ApoB region encompassing the editable site. (A) Schematic diagram of the chimeric transcript. The mCherry cds is placed in frame 5′ to the EGFP cds. The region of ApoB encompassing the C6666 editable site links the 2 coding sequences. APOBEC1-specific RNA editing induces the formation of a stop codon that blocks the translation of the EGFP coding sequence. The mCherry protein acts as a control for the transfection levels, while the loss of EGFP is indicative of the levels of RNA editing. The boxes on the side of the editable site indicate the efficiency and the mooring sequence (labeled e and m, respectively). (B) Representative FACS analysis of HuH-7 cells transiently transfected with the mCherry-ApoB-EGFP construct together with those expressing respectively rat APOBEC1, a catalytically inactive APOBEC1 mutant (E63A), a mutant lacking the C-terminal 40aa (ΔC), or control plasmids. Only rat APOBEC1 induces a shift from the mCherry/EGFP diagonal (events in the red-boxed area), indicating an overall decrease in the EGFP levels. The gates selected for the analysis, as well as ungated plots are shown in Supplementary Figure 1. (C) Representative chromatograms of the ApoB editing site of the mCherry-ApoB-EGFP transcript from cells transfected either with control plasmids or with the APOBEC1 expressing one. In the APOBEC1 sample the chromatogram shows a double peak corresponding to the C>U deamination (arrow).
