Fig. 3.
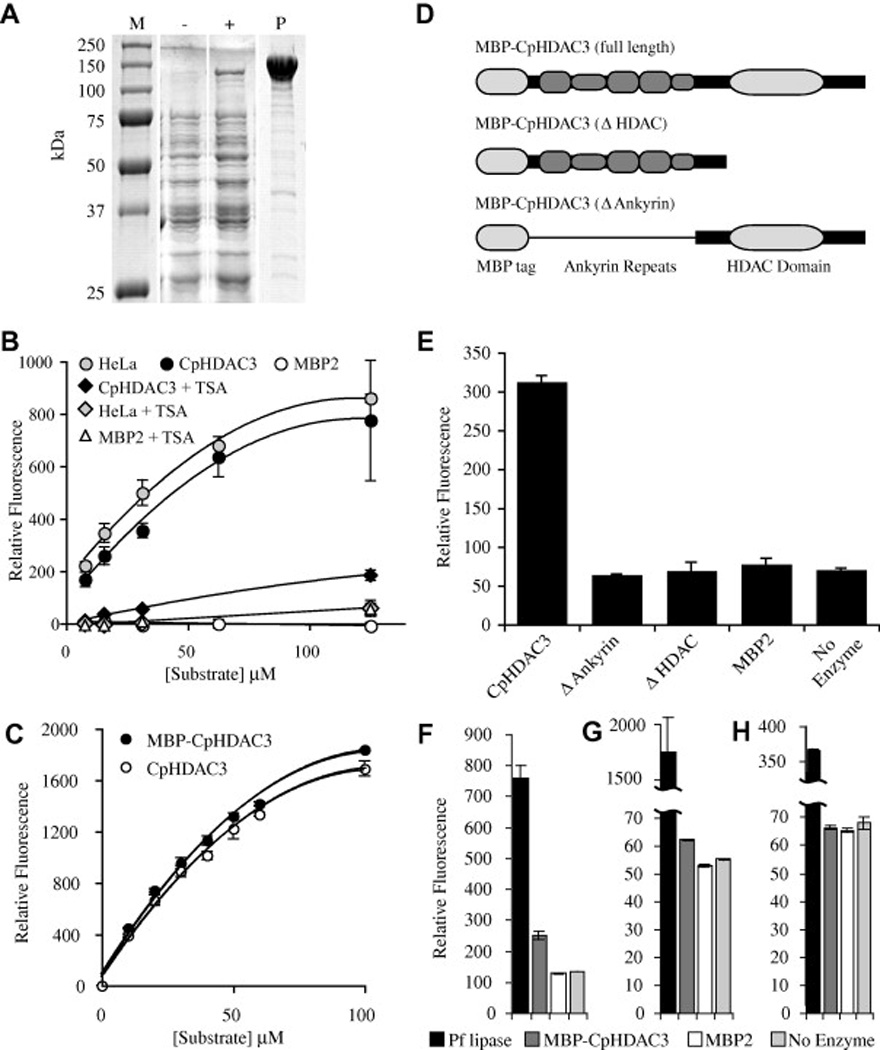
Activity of recombinant Cryptosporidium parvum histone deacetylase 3 (CpHDAC3). (A) Protein gel of recombinant CpHDAC3. Lane M contains protein marker with sizes indicated to the left of the figure. Whole cell lysates are presented for bacteria harbouring an expression plasmid encoding a maltose binding protein fusion to CpHDAC3 before (−) and after (+) induction with isopropyl β-d-1-thiogalactopyranoside. Lane P contains affinity-purified recombinant maltose binding protein (MBP)-CpHDAC3. (B) In vitro assays with a fluorogenic HDAC substrate were compared with human HeLa cell nuclear extract as a positive control and MBP2 as a negative control. With the broad-spectrum HDAC inhibitor trichostatin A, CpHDAC3 was suppressible to levels near that of a no-enzyme control. (C) Comparison of equimolar amounts of MBP-CpHDAC3 fusion (uncut) and CpHDAC3 (cut) indicated equivalent activities for both proteins. Kinetic data are presented in the main text. (D) Schematic representations of full-length and truncated MBP fusions of CpHDAC3. A thin line running through the N-terminal truncation is to indicate that the MBP is linked to the C-terminal region containing only the HDAC domain. (E) HDAC Activity of MBP-CpHDAC3 and truncated versions together with MBP2 and no-enzyme negative controls. For simplicity, only a single concentration of substrate is presented (10 µM). (F–H) In vitro assays with (F) 250 µM 4-methylumbelliferyl acetate, (G) 4-methylumbelliferyl propionate or (H) 4-methylumbelliferyl butyrate as esterase substrates compared with Pseudomonas fluorescens lipase (Pf lipase) as a positive control. MBP-CpHDAC3 displayed activity against acetyl and propionyl esters (but not butyryl esters) that was significantly higher than MBP2 or no-enzyme controls. Kinetic data are presented in Table 1.
