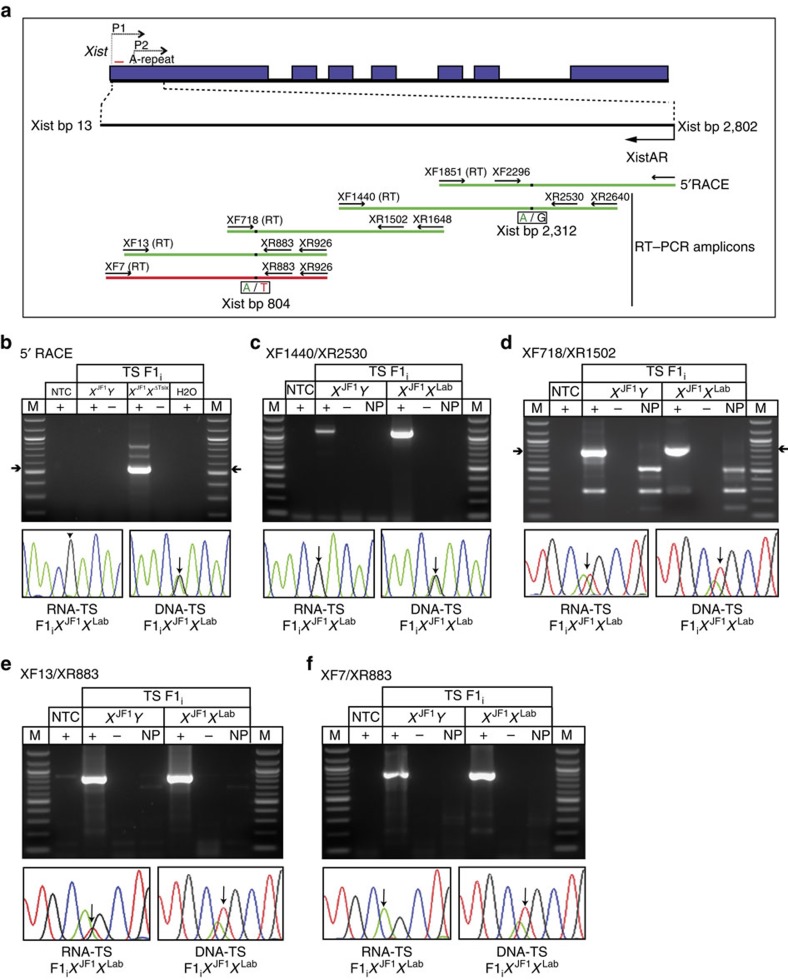
Figure 3. Mapping of XistAR.
(a) Schematic representation of 5′ RACE and RT–PCR strategy to delineate the structure of XistAR in WT F1i hybrid TS cells. SNPs that distinguish the maternal and paternal X-chromosome alleles (Xm and Xp, respectively) are shown under the amplicons. Green amplicons, detection of XistAR expression from the inactive X. Red amplicon, lack of XistAR detection. (b) Mapping of 5′ end of XistAR to bp 2,802 of Xist by 5′ RLM-RACE. Sanger sequencing of the major amplified cDNA detects Xp-specific SNPs. Amplified genomic DNA displays SNPs from both alleles. (c–f) Mapping of XistAR by overlapping RT–PCRs. All amplicons except XF7/XR883 detect XistAR. The 3′ end of XistAR therefore maps to ∼Xist bp 13. In c Sanger sequencing demonstrates primary amplification of XistAR, but not the Xist antisense transcript Tsix. The PCR primers XR2640 and XR2530 in this amplicon map to intron 3 of Tsix, and thus do not amplify spliced Tsix RNA. The low-level amplification in males is presumptive unspliced Tsix RNA. In d,e Sanger sequencing detects SNPs from both Xs, since both XistAR and Tsix are reverse transcribed and amplified. In f, Sanger sequencing detects amplification of Tsix but not XistAR. M, marker; NTC, no template control; +, reaction with reverse transcriptase (RT); −, reaction without RT; NP, ‘no primer' control RT reaction without added primers.