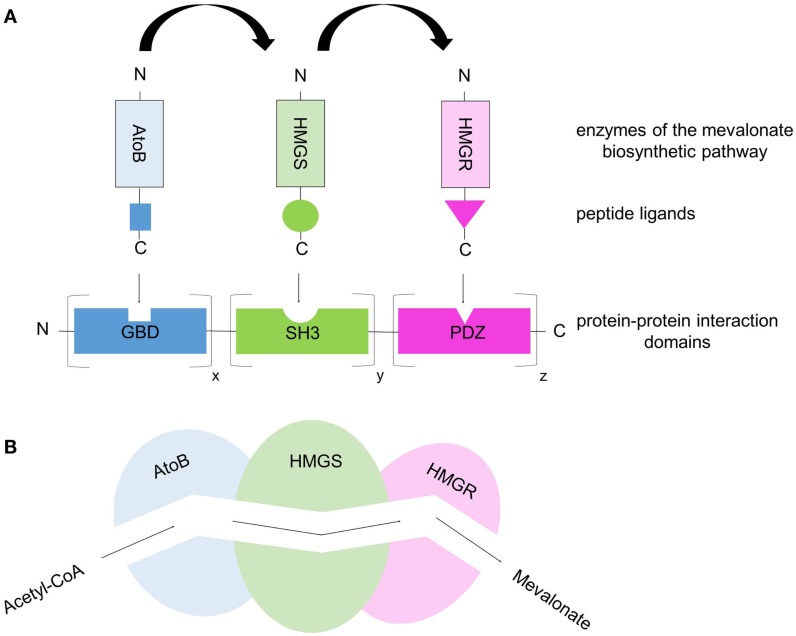
Figure 4.
Protein-scaffold-based multienzyme complex offers a synthetic metabolic pipeline that allows substrate channeling. (A) The three enzymes of the mevalonate biosynthetic pathway (acetoacetyl-CoA thiolase, AtoB; hydroxy-methylglutaryl-CoA synthase, HMGS; hydroxy-methylglutaryl-CoA reductase, HMGR) were co-localized on a synthetic protein-scaffold via high-affinity interactions between protein domains and their specific, cognate peptide ligands. The protein scaffold consists of various repeats of well-characterized, metazoan protein domains (SH3, GBD, PDZ) and the corresponding peptide ligands are fused to the enzymes. By co-expressing the synthetic scaffold and the enzyme–peptide ligand fusions in E.coli cells, the three enzymes are targeted to the scaffold building a multienzyme complex. Through the variation of the binding domain repeats the ratio/stoichiometry of enzymes can be controlled. (B) Simplified scheme of substrate channeling through a tunnel/metabolic pipeline connecting the active sites of the co-localized enzymes.