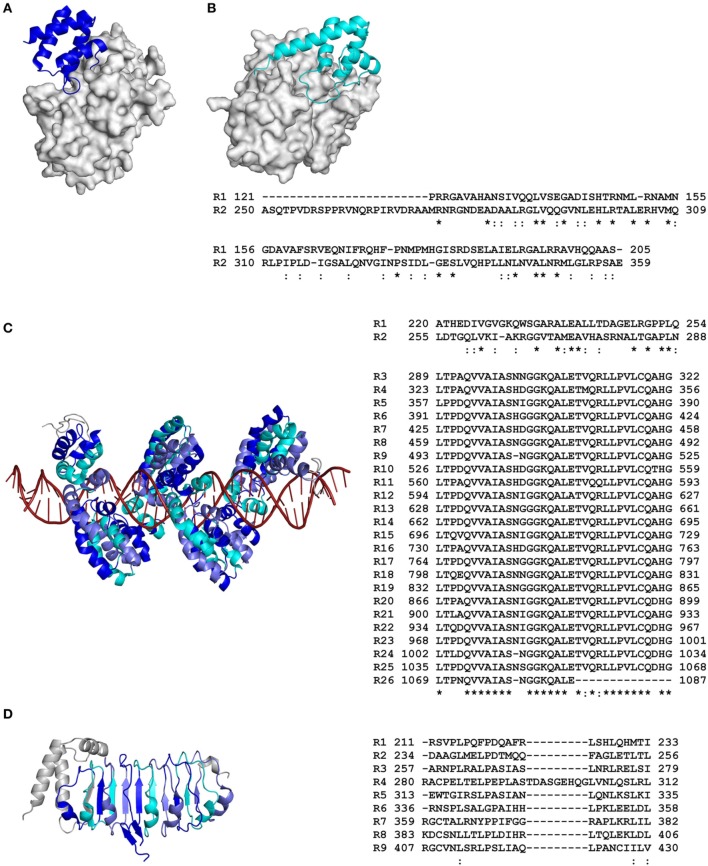
Figure 1.
Primary and tertiary structures of repeat domains from RCP effectors of plant-associated bacteria. (A) Crystal structure of repeat unit one from the AvrPtoB effector of the tomato bacterial speck pathogen, Pseudomonas syringae pv. tomato (Pst), in complex with the tomato Pto kinase (Protein Data Bank [PDB] code 3HGK; Dong et al., 2009). (B) Nuclear magnetic resonance (NMR) structure of repeat unit two from AvrPtoB of Pst in complex with the BAK1 kinase domain from Arabidopsis thaliana (3TL8; Cheng et al., 2011). Note that in (A), AvrPtoB repeat unit one interacts with the Pto kinase in a different orientation to that of AvrPtoB repeat unit two with the BAK1 kinase domain in (B). (C) Crystal structure of the repeat domain from the PthXo1 transcription activator-like (TAL) effector of the bacterial rice pathogen, Xanthomonas oryzae pv. oryzae, bound to its natural DNA target (36 bp). The repeats pack together to form a left-handed superhelix (α-solenoid) that wraps around the DNA molecule (3UGM; Mak et al., 2012). (D) Crystal structure of the N-terminal leucine-rich repeat (LRR) domain from the XopL effector of the bacterial leaf spot pathogen of pepper and tomato, Xanthomonas euvesicatoria (4FCG; Singer et al., 2013). Structural coordinate files were downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) PDB (http://www.rcsb.org/pdb/home/home.do). Alternating repeat units are colored blue, slate, and cyan, respectively. Non-repetitive sequence is colored gray. The molecular surface of Pto kinase in (A) and BAK1 kinase domain in (B) are shown in gray, while the DNA molecule in (C) is colored red. An amino acid sequence alignment detailing the primary structure of each RCP effector repeat domain is shown to the right of each tertiary structure (as based on that presented in each tertiary structure). Repeat (R) units are numbered according to their position in the RCP effector. The start and end position of each repeat unit in the full-length RCP effector is shown. Conserved (*) and strongly similar (:) amino acid residues shared between repeat units are shown below the sequence alignment (based on full-length repeat units only). The figure was prepared using PyMol (https://www.pymol.org/) and Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/).